
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 23079975 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| SKCM | ||
| COAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000364677 | zf-C2H2 | PF00096.26 | 3.2e-13 | 1 | 2 |
| ENSP00000364677 | zf-C2H2 | PF00096.26 | 3.2e-13 | 2 | 2 |
| ENSP00000364677 | zf-met | PF12874.7 | 0.00019 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000375527 | ZBTB12-201 | 1875 | XM_011514383 | ENSP00000364677 | 459 (aa) | XP_011512685 | Q9Y330 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000204366 | Lupus Erythematosus, Systemic | 2.8947E-18 | 18204098 |
| ENSG00000204366 | Diabetes Mellitus, Type 1 | 1.6E-86 | 19430480 |
| ENSG00000204366 | Lupus Erythematosus, Systemic | 4.2E-8 | 18204446 |
| ENSG00000204366 | Lupus Erythematosus, Systemic | 8E-21 | 24871463 |
| ENSG00000204366 | Vaccine Potency | 1E-17 | 21764829 |
| ENSG00000204366 | Surveys and Questionnaires | 6E-6 | 22566634 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit increase | 1.5771569 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit increase | 1.1050375 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit increase | 0.89926857 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit decrease | 0.5455914 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit decrease | 0.51218635 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [NR] unit increase | 0.45406085 | EFO_0007937 |
| ENSG00000204366 | rs114508013 | 6 | 31900988 | G | Blood protein levels | 30072576 | [1.04-1.31] unit increase | 1.176 | EFO_0007937 |
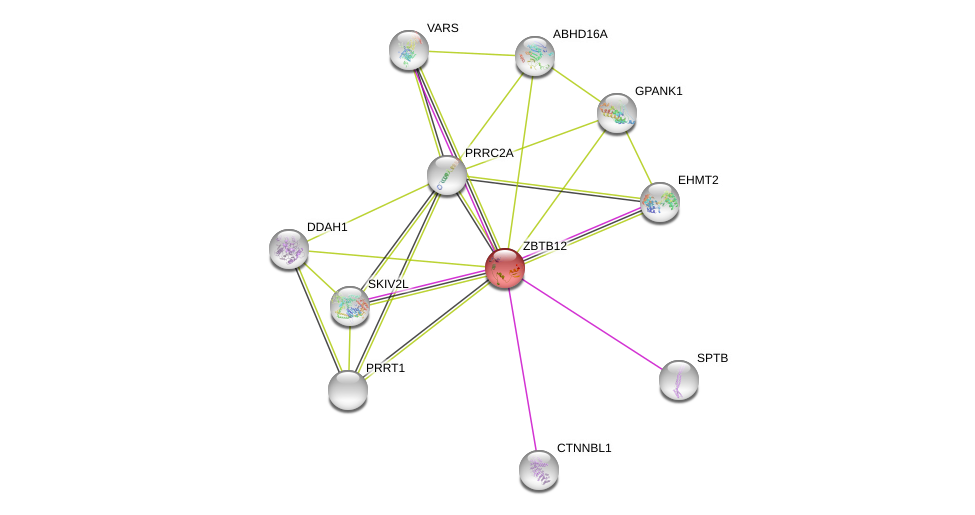
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000204366 | ZBTB12 | 100 | 96.296 | ENSAMEG00000012921 | ZBTB12 | 100 | 96.296 | Ailuropoda_melanoleuca |
| ENSG00000204366 | ZBTB12 | 100 | 64.571 | ENSACAG00000011189 | ZBTB12 | 100 | 64.397 | Anolis_carolinensis |
| ENSG00000204366 | ZBTB12 | 100 | 99.782 | ENSANAG00000019564 | ZBTB12 | 100 | 99.782 | Aotus_nancymaae |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSBTAG00000032031 | ZBTB12 | 100 | 98.911 | Bos_taurus |
| ENSG00000204366 | ZBTB12 | 100 | 99.564 | ENSCJAG00000044185 | ZBTB12 | 100 | 99.564 | Callithrix_jacchus |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSCAFG00000000674 | ZBTB12 | 100 | 98.475 | Canis_familiaris |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSCAFG00020000911 | ZBTB12 | 100 | 98.475 | Canis_lupus_dingo |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSCHIG00000007636 | ZBTB12 | 100 | 98.911 | Capra_hircus |
| ENSG00000204366 | ZBTB12 | 100 | 99.346 | ENSCCAG00000019304 | ZBTB12 | 100 | 99.346 | Cebus_capucinus |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSCLAG00000007108 | ZBTB12 | 100 | 98.911 | Chinchilla_lanigera |
| ENSG00000204366 | ZBTB12 | 99 | 65.605 | ENSCPBG00000012202 | ZBTB12 | 99 | 67.797 | Chrysemys_picta_bellii |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSCANG00000038177 | ZBTB12 | 100 | 98.475 | Colobus_angolensis_palliatus |
| ENSG00000204366 | ZBTB12 | 100 | 95.474 | ENSCGRG00001002052 | Zbtb12 | 100 | 95.474 | Cricetulus_griseus_chok1gshd |
| ENSG00000204366 | ZBTB12 | 53 | 67.717 | ENSDARG00000103949 | zbtb12.1 | 51 | 67.717 | Danio_rerio |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSEASG00005015654 | ZBTB12 | 100 | 98.475 | Equus_asinus_asinus |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSECAG00000034980 | ZBTB12 | 100 | 98.475 | Equus_caballus |
| ENSG00000204366 | ZBTB12 | 100 | 98.693 | ENSFCAG00000031296 | ZBTB12 | 100 | 98.693 | Felis_catus |
| ENSG00000204366 | ZBTB12 | 77 | 95.122 | ENSFDAG00000018453 | ZBTB12 | 99 | 96.855 | Fukomys_damarensis |
| ENSG00000204366 | ZBTB12 | 99 | 65.678 | ENSGAGG00000002344 | ZBTB12 | 99 | 67.511 | Gopherus_agassizii |
| ENSG00000204366 | ZBTB12 | 100 | 99.346 | ENSGGOG00000015638 | ZBTB12 | 100 | 99.346 | Gorilla_gorilla |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSHGLG00000016731 | ZBTB12 | 100 | 98.911 | Heterocephalus_glaber_female |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSSTOG00000027411 | ZBTB12 | 100 | 98.911 | Ictidomys_tridecemlineatus |
| ENSG00000204366 | ZBTB12 | 99 | 60.752 | ENSLACG00000006007 | ZBTB12 | 99 | 60.970 | Latimeria_chalumnae |
| ENSG00000204366 | ZBTB12 | 100 | 99.782 | ENSMFAG00000010877 | ZBTB12 | 100 | 99.782 | Macaca_fascicularis |
| ENSG00000204366 | ZBTB12 | 100 | 99.782 | ENSMMUG00000011683 | ZBTB12 | 100 | 99.782 | Macaca_mulatta |
| ENSG00000204366 | ZBTB12 | 100 | 99.782 | ENSMNEG00000022456 | ZBTB12 | 100 | 99.782 | Macaca_nemestrina |
| ENSG00000204366 | ZBTB12 | 100 | 96.739 | ENSMAUG00000008051 | Zbtb12 | 100 | 96.739 | Mesocricetus_auratus |
| ENSG00000204366 | ZBTB12 | 100 | 99.129 | ENSMICG00000003600 | ZBTB12 | 100 | 99.129 | Microcebus_murinus |
| ENSG00000204366 | ZBTB12 | 100 | 97.168 | ENSMOCG00000002711 | Zbtb12 | 100 | 97.168 | Microtus_ochrogaster |
| ENSG00000204366 | ZBTB12 | 99 | 81.761 | ENSMODG00000015096 | - | 96 | 81.132 | Monodelphis_domestica |
| ENSG00000204366 | ZBTB12 | 100 | 97.386 | MGP_CAROLIEiJ_G0021482 | Zbtb12 | 100 | 97.386 | Mus_caroli |
| ENSG00000204366 | ZBTB12 | 100 | 97.386 | ENSMUSG00000049823 | Zbtb12 | 100 | 97.386 | Mus_musculus |
| ENSG00000204366 | ZBTB12 | 100 | 97.168 | MGP_PahariEiJ_G0020477 | Zbtb12 | 100 | 97.168 | Mus_pahari |
| ENSG00000204366 | ZBTB12 | 100 | 97.386 | MGP_SPRETEiJ_G0022387 | Zbtb12 | 100 | 97.386 | Mus_spretus |
| ENSG00000204366 | ZBTB12 | 100 | 98.911 | ENSMPUG00000018357 | ZBTB12 | 100 | 98.911 | Mustela_putorius_furo |
| ENSG00000204366 | ZBTB12 | 100 | 96.514 | ENSNGAG00000011842 | Zbtb12 | 100 | 96.514 | Nannospalax_galili |
| ENSG00000204366 | ZBTB12 | 100 | 99.782 | ENSNLEG00000034429 | ZBTB12 | 100 | 99.782 | Nomascus_leucogenys |
| ENSG00000204366 | ZBTB12 | 100 | 97.821 | ENSODEG00000017709 | ZBTB12 | 100 | 97.821 | Octodon_degus |
| ENSG00000204366 | ZBTB12 | 100 | 99.346 | ENSOGAG00000031042 | ZBTB12 | 100 | 99.346 | Otolemur_garnettii |
| ENSG00000204366 | ZBTB12 | 100 | 94.553 | ENSPPAG00000033581 | ZBTB12 | 100 | 95.207 | Pan_paniscus |
| ENSG00000204366 | ZBTB12 | 100 | 98.693 | ENSPPRG00000000449 | ZBTB12 | 100 | 98.693 | Panthera_pardus |
| ENSG00000204366 | ZBTB12 | 100 | 99.564 | ENSPTRG00000017993 | ZBTB12 | 100 | 99.564 | Pan_troglodytes |
| ENSG00000204366 | ZBTB12 | 100 | 99.564 | ENSPANG00000003484 | ZBTB12 | 100 | 99.564 | Papio_anubis |
| ENSG00000204366 | ZBTB12 | 64 | 99.658 | ENSPPYG00000016468 | - | 100 | 99.658 | Pongo_abelii |
| ENSG00000204366 | ZBTB12 | 100 | 99.346 | ENSPCOG00000013160 | ZBTB12 | 100 | 99.346 | Propithecus_coquereli |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSPVAG00000005130 | ZBTB12 | 100 | 98.475 | Pteropus_vampyrus |
| ENSG00000204366 | ZBTB12 | 100 | 97.821 | ENSRNOG00000000418 | Zbtb12 | 100 | 97.821 | Rattus_norvegicus |
| ENSG00000204366 | ZBTB12 | 99 | 74.890 | ENSSHAG00000008383 | ZBTB12 | 97 | 72.907 | Sarcophilus_harrisii |
| ENSG00000204366 | ZBTB12 | 94 | 75.221 | ENSSPUG00000013016 | ZBTB12 | 82 | 75.325 | Sphenodon_punctatus |
| ENSG00000204366 | ZBTB12 | 100 | 99.129 | ENSSSCG00000001421 | ZBTB12 | 100 | 99.129 | Sus_scrofa |
| ENSG00000204366 | ZBTB12 | 100 | 97.216 | ENSUAMG00000024479 | ZBTB12 | 100 | 97.216 | Ursus_americanus |
| ENSG00000204366 | ZBTB12 | 100 | 98.475 | ENSVVUG00000022537 | ZBTB12 | 100 | 98.475 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |