
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| HNSC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| ACC | |||
| CHOL | |||
| HNSC | |||
| KIRP | |||
| LUSC | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| ESCA | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000336762 | RnaseA | PF00074.20 | 1e-34 | 1 | 1 |
| ENSP00000381077 | RnaseA | PF00074.20 | 1e-34 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000336811 | ANG-201 | 1222 | - | ENSP00000336762 | 147 (aa) | - | P03950 |
| ENST00000397990 | ANG-202 | 742 | - | ENSP00000381077 | 147 (aa) | - | P03950 |
| ENST00000554073 | ANG-203 | 251 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000214274 | rs184297073 | 14 | 20690905 | T | Blood protein levels | 29875488 | [1.78-2.44] unit decrease | 2.11 | EFO_0007937 |
| ENSG00000214274 | rs1888562 | 14 | 20696377 | T | Blood protein levels | 29875488 | [0.23-0.35] unit decrease | 0.29 | EFO_0007937 |
| ENSG00000214274 | rs17114671 | 14 | 20687111 | C | Blood protein levels | 28240269 | [0.69-0.92] unit increase | 0.8064 | EFO_0008022 |
| ENSG00000214274 | rs1888560 | 14 | 20696086 | T | Blood protein levels | 28240269 | [0.26-0.43] unit decrease | 0.3446 | EFO_0008022 |
| ENSG00000214274 | rs17114699 | 14 | 20690751 | T | Blood protein levels | 28240269 | [0.63-0.88] unit increase | 0.7532 | EFO_0008022 |
| ENSG00000214274 | rs1888560 | 14 | 20696086 | C | Blood protein levels | 30072576 | [0.17-0.27] unit increase | 0.223 | EFO_0007937 |
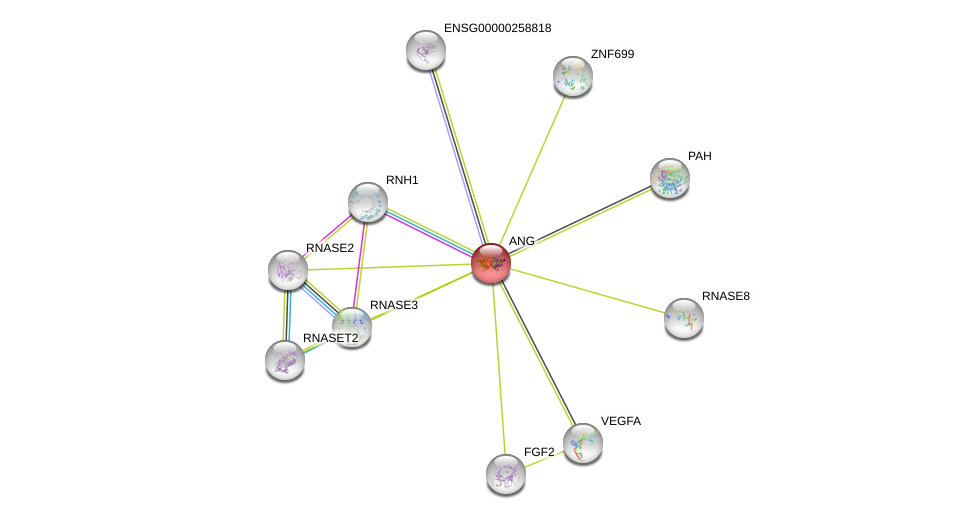
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000214274 | ANG | 82 | 39.837 | ENSG00000258818 | RNASE4 | 83 | 39.837 |
| ENSG00000214274 | ANG | 76 | 32.283 | ENSG00000165799 | RNASE7 | 76 | 32.283 |
| ENSG00000214274 | ANG | 86 | 35.507 | ENSG00000129538 | RNASE1 | 94 | 36.522 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000214274 | ANG | 99 | 77.241 | ENSAMEG00000018660 | ANG | 87 | 76.378 | Ailuropoda_melanoleuca |
| ENSG00000214274 | ANG | 93 | 63.504 | ENSBTAG00000032873 | - | 72 | 61.983 | Bos_taurus |
| ENSG00000214274 | ANG | 97 | 66.434 | ENSBTAG00000019612 | RNASE4 | 97 | 65.972 | Bos_taurus |
| ENSG00000214274 | ANG | 98 | 55.556 | ENSBTAG00000046668 | ANG2 | 97 | 55.556 | Bos_taurus |
| ENSG00000214274 | ANG | 97 | 64.336 | ENSCHIG00000010811 | - | 97 | 64.336 | Capra_hircus |
| ENSG00000214274 | ANG | 97 | 66.434 | ENSCHIG00000010308 | - | 97 | 65.972 | Capra_hircus |
| ENSG00000214274 | ANG | 78 | 48.462 | ENSCPBG00000000657 | - | 82 | 48.462 | Chrysemys_picta_bellii |
| ENSG00000214274 | ANG | 85 | 42.063 | ENSCPBG00000009437 | - | 85 | 41.600 | Chrysemys_picta_bellii |
| ENSG00000214274 | ANG | 98 | 67.361 | ENSCGRG00001006777 | - | 58 | 68.000 | Cricetulus_griseus_chok1gshd |
| ENSG00000214274 | ANG | 98 | 67.361 | ENSCGRG00000016447 | - | 79 | 68.000 | Cricetulus_griseus_crigri |
| ENSG00000214274 | ANG | 99 | 75.172 | ENSEASG00005002967 | ANG | 87 | 74.803 | Equus_asinus_asinus |
| ENSG00000214274 | ANG | 99 | 75.172 | ENSECAG00000035254 | ANG | 87 | 74.803 | Equus_caballus |
| ENSG00000214274 | ANG | 99 | 78.621 | ENSFCAG00000041462 | ANG | 99 | 78.621 | Felis_catus |
| ENSG00000214274 | ANG | 79 | 48.855 | ENSGAGG00000006587 | - | 64 | 48.855 | Gopherus_agassizii |
| ENSG00000214274 | ANG | 92 | 42.647 | ENSGAGG00000019734 | - | 85 | 41.600 | Gopherus_agassizii |
| ENSG00000214274 | ANG | 100 | 99.320 | ENSGGOG00000044112 | ANG | 100 | 99.320 | Gorilla_gorilla |
| ENSG00000214274 | ANG | 98 | 67.361 | ENSMAUG00000010475 | - | 86 | 68.000 | Mesocricetus_auratus |
| ENSG00000214274 | ANG | 99 | 67.808 | ENSMOCG00000000621 | - | 88 | 68.750 | Microtus_ochrogaster |
| ENSG00000214274 | ANG | 87 | 63.077 | ENSMODG00000006972 | ANG | 81 | 63.077 | Monodelphis_domestica |
| ENSG00000214274 | ANG | 98 | 74.306 | ENSMUSG00000072115 | Ang | 86 | 78.400 | Mus_musculus |
| ENSG00000214274 | ANG | 97 | 66.434 | ENSMUSG00000047894 | Ang2 | 85 | 68.548 | Mus_musculus |
| ENSG00000214274 | ANG | 84 | 66.129 | ENSMUSG00000060615 | Ang4 | 85 | 66.129 | Mus_musculus |
| ENSG00000214274 | ANG | 97 | 65.035 | ENSMUSG00000053961 | Ang5 | 85 | 66.935 | Mus_musculus |
| ENSG00000214274 | ANG | 97 | 57.241 | ENSMUSG00000072598 | Ang6 | 98 | 57.241 | Mus_musculus |
| ENSG00000214274 | ANG | 98 | 70.833 | MGP_PahariEiJ_G0030242 | - | 86 | 72.800 | Mus_pahari |
| ENSG00000214274 | ANG | 98 | 70.833 | MGP_PahariEiJ_G0030240 | - | 86 | 72.800 | Mus_pahari |
| ENSG00000214274 | ANG | 98 | 73.611 | MGP_SPRETEiJ_G0020109 | Ang | 86 | 77.600 | Mus_spretus |
| ENSG00000214274 | ANG | 98 | 65.278 | MGP_SPRETEiJ_G0021593 | - | 86 | 67.200 | Mus_spretus |
| ENSG00000214274 | ANG | 98 | 65.278 | MGP_SPRETEiJ_G0021592 | - | 86 | 67.200 | Mus_spretus |
| ENSG00000214274 | ANG | 99 | 68.966 | ENSMLUG00000024437 | - | 87 | 69.291 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 99 | 68.276 | ENSMLUG00000025641 | - | 87 | 68.504 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 87 | 68.750 | ENSMLUG00000029656 | - | 87 | 68.504 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 98 | 68.750 | ENSMLUG00000029597 | - | 88 | 69.048 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 98 | 65.278 | ENSMLUG00000027892 | - | 86 | 65.873 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 98 | 68.750 | ENSMLUG00000028700 | - | 88 | 69.048 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 86 | 71.654 | ENSMLUG00000027018 | - | 85 | 71.429 | Myotis_lucifugus |
| ENSG00000214274 | ANG | 98 | 78.472 | ENSNGAG00000018469 | - | 86 | 80.952 | Nannospalax_galili |
| ENSG00000214274 | ANG | 95 | 61.538 | ENSMEUG00000000681 | ANG | 83 | 62.712 | Notamacropus_eugenii |
| ENSG00000214274 | ANG | 98 | 63.194 | ENSOGAG00000027209 | ANG | 86 | 61.111 | Otolemur_garnettii |
| ENSG00000214274 | ANG | 98 | 54.861 | ENSOARG00000013005 | - | 97 | 54.861 | Ovis_aries |
| ENSG00000214274 | ANG | 97 | 63.636 | ENSOARG00000012562 | - | 97 | 63.636 | Ovis_aries |
| ENSG00000214274 | ANG | 100 | 98.639 | ENSPPAG00000032810 | ANG | 100 | 98.639 | Pan_paniscus |
| ENSG00000214274 | ANG | 99 | 78.621 | ENSPPRG00000020077 | ANG | 99 | 78.621 | Panthera_pardus |
| ENSG00000214274 | ANG | 99 | 78.621 | ENSPTIG00000004149 | ANG | 99 | 78.621 | Panthera_tigris_altaica |
| ENSG00000214274 | ANG | 100 | 99.320 | ENSPTRG00000043768 | ANG | 100 | 99.320 | Pan_troglodytes |
| ENSG00000214274 | ANG | 94 | 41.892 | ENSPKIG00000000354 | - | 66 | 44.800 | Paramormyrops_kingsleyae |
| ENSG00000214274 | ANG | 80 | 43.939 | ENSPSIG00000000732 | ANG | 82 | 43.939 | Pelodiscus_sinensis |
| ENSG00000214274 | ANG | 98 | 69.444 | ENSPEMG00000013152 | - | 86 | 69.048 | Peromyscus_maniculatus_bairdii |
| ENSG00000214274 | ANG | 97 | 54.545 | ENSPCAG00000006037 | ANG | 84 | 52.000 | Procavia_capensis |
| ENSG00000214274 | ANG | 99 | 78.621 | ENSPCOG00000013248 | ANG | 99 | 78.621 | Propithecus_coquereli |
| ENSG00000214274 | ANG | 99 | 65.517 | ENSPVAG00000013562 | ANG | 99 | 65.517 | Pteropus_vampyrus |
| ENSG00000214274 | ANG | 86 | 74.603 | ENSRNOG00000025562 | Ang2 | 75 | 74.603 | Rattus_norvegicus |
| ENSG00000214274 | ANG | 86 | 74.603 | ENSRNOG00000043032 | Ang2 | 75 | 74.603 | Rattus_norvegicus |
| ENSG00000214274 | ANG | 93 | 56.081 | ENSSHAG00000006482 | ANG | 82 | 63.200 | Sarcophilus_harrisii |
| ENSG00000214274 | ANG | 98 | 70.139 | ENSSSCG00000031538 | ANG | 80 | 69.048 | Sus_scrofa |
| ENSG00000214274 | ANG | 97 | 65.734 | ENSTTRG00000009493 | ANG | 99 | 65.734 | Tursiops_truncatus |
| ENSG00000214274 | ANG | 99 | 76.552 | ENSUAMG00000012125 | ANG | 87 | 76.378 | Ursus_americanus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001525 | angiogenesis | 8448182. | IDA | Process |
| GO:0001525 | angiogenesis | 2479414.17125737. | IMP | Process |
| GO:0001525 | angiogenesis | 16567967. | TAS | Process |
| GO:0001541 | ovarian follicle development | 12770725. | NAS | Process |
| GO:0001556 | oocyte maturation | 11438326. | NAS | Process |
| GO:0001666 | response to hypoxia | 10999833.15979542.16490744. | IDA | Process |
| GO:0001666 | response to hypoxia | 15776477. | NAS | Process |
| GO:0001890 | placenta development | 11984825. | NAS | Process |
| GO:0001938 | positive regulation of endothelial cell proliferation | 9122172.9707554. | IDA | Process |
| GO:0003677 | DNA binding | 10649442. | IC | Function |
| GO:0003779 | actin binding | 7679494.11782452. | IDA | Function |
| GO:0004519 | endonuclease activity | 10103013. | TAS | Function |
| GO:0004540 | ribonuclease activity | 21873635. | IBA | Function |
| GO:0004540 | ribonuclease activity | 2424496.2730651. | IDA | Function |
| GO:0005102 | signaling receptor binding | 9122172. | IDA | Function |
| GO:0005507 | copper ion binding | 9245697. | IDA | Function |
| GO:0005515 | protein binding | 2742853.3470787.10413501.15737636.17991437.24457100.28777577. | IPI | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005604 | basement membrane | 15166501. | IDA | Component |
| GO:0005615 | extracellular space | 3663649.16461950.16490744. | IDA | Component |
| GO:0005634 | nucleus | 10649442.15735021.25372031. | IDA | Component |
| GO:0005730 | nucleolus | - | ISS | Component |
| GO:0006651 | diacylglycerol biosynthetic process | 2457905. | IDA | Process |
| GO:0007154 | cell communication | 10103013. | NAS | Process |
| GO:0007202 | activation of phospholipase C activity | 2457905. | IMP | Process |
| GO:0008201 | heparin binding | 10103013. | IDA | Function |
| GO:0009303 | rRNA transcription | 15735021. | IMP | Process |
| GO:0009725 | response to hormone | 10999833. | IDA | Process |
| GO:0016477 | cell migration | 17125737. | IMP | Process |
| GO:0017148 | negative regulation of translation | - | IEA | Process |
| GO:0019843 | rRNA binding | 2457905. | TAS | Function |
| GO:0030041 | actin filament polymerization | - | ISS | Process |
| GO:0030426 | growth cone | - | ISS | Component |
| GO:0031410 | cytoplasmic vesicle | - | IEA | Component |
| GO:0032148 | activation of protein kinase B activity | 17125737. | IMP | Process |
| GO:0032311 | angiogenin-PRI complex | 3470787. | IDA | Component |
| GO:0032311 | angiogenin-PRI complex | 3470787. | IPI | Component |
| GO:0032431 | activation of phospholipase A2 activity | 2646638. | IMP | Process |
| GO:0034332 | adherens junction organization | - | TAS | Process |
| GO:0042277 | peptide binding | 11782452. | IDA | Function |
| GO:0042327 | positive regulation of phosphorylation | 17125737. | IDA | Process |
| GO:0042592 | homeostatic process | 15166501. | NAS | Process |
| GO:0042803 | protein homodimerization activity | 25372031. | IDA | Function |
| GO:0043025 | neuronal cell body | - | ISS | Component |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | 10103013. | IDA | Process |
| GO:0050714 | positive regulation of protein secretion | 2646638. | IDA | Process |
| GO:0050714 | positive regulation of protein secretion | - | ISS | Process |
| GO:0090501 | RNA phosphodiester bond hydrolysis | - | IEA | Process |