
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| DLBC | |||
| ESCA | |||
| PAAD | |||
| READ | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| LAML |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000333329 | HABP4_PAI-RBP1 | PF04774.15 | 0.0014 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 17109758 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000333796 | USP17L2-201 | 1910 | - | ENSP00000333329 | 530 (aa) | - | Q6R6M4 |
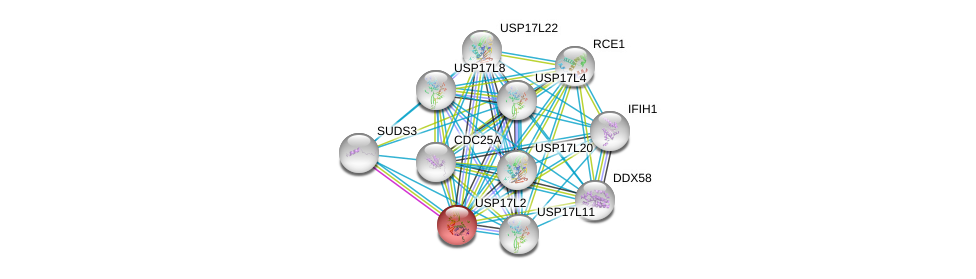
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000223443 | USP17L2 | 100 | 94.340 | ENSGGOG00000042639 | - | 100 | 94.340 | Gorilla_gorilla |
| ENSG00000223443 | USP17L2 | 74 | 95.475 | ENSGGOG00000009925 | - | 94 | 95.475 | Gorilla_gorilla |
| ENSG00000223443 | USP17L2 | 100 | 87.170 | ENSGGOG00000043616 | - | 100 | 87.170 | Gorilla_gorilla |
| ENSG00000223443 | USP17L2 | 78 | 94.712 | ENSGGOG00000014367 | - | 100 | 94.712 | Gorilla_gorilla |
| ENSG00000223443 | USP17L2 | 100 | 94.340 | ENSGGOG00000044031 | - | 100 | 94.340 | Gorilla_gorilla |
| ENSG00000223443 | USP17L2 | 100 | 88.491 | ENSMFAG00000043645 | - | 95 | 88.491 | Macaca_fascicularis |
| ENSG00000223443 | USP17L2 | 87 | 88.961 | ENSMFAG00000039016 | - | 100 | 88.961 | Macaca_fascicularis |
| ENSG00000223443 | USP17L2 | 100 | 88.491 | ENSMFAG00000036389 | - | 95 | 88.491 | Macaca_fascicularis |
| ENSG00000223443 | USP17L2 | 100 | 88.302 | ENSMMUG00000049137 | - | 100 | 88.302 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.302 | ENSMMUG00000040484 | - | 100 | 88.302 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.491 | ENSMMUG00000047519 | - | 100 | 88.491 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 86.604 | ENSMMUG00000039802 | - | 100 | 86.604 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 89.057 | ENSMMUG00000041878 | - | 100 | 89.057 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 81.887 | ENSMMUG00000044213 | - | 100 | 81.887 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.113 | ENSMMUG00000041488 | - | 100 | 88.113 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 99 | 87.689 | ENSMMUG00000045288 | - | 95 | 87.689 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 83.774 | ENSMMUG00000042380 | - | 96 | 83.774 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.113 | ENSMMUG00000049096 | - | 96 | 88.113 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.113 | ENSMMUG00000042715 | - | 100 | 88.113 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 100 | 88.302 | ENSMMUG00000029040 | - | 96 | 88.302 | Macaca_mulatta |
| ENSG00000223443 | USP17L2 | 99 | 79.735 | ENSMNEG00000033266 | - | 96 | 80.114 | Macaca_nemestrina |
| ENSG00000223443 | USP17L2 | 100 | 77.444 | ENSMLEG00000029099 | - | 100 | 77.444 | Mandrillus_leucophaeus |
| ENSG00000223443 | USP17L2 | 100 | 92.075 | ENSNLEG00000035477 | - | 100 | 92.075 | Nomascus_leucogenys |
| ENSG00000223443 | USP17L2 | 100 | 84.717 | ENSPTRG00000049794 | - | 100 | 84.717 | Pan_troglodytes |
| ENSG00000223443 | USP17L2 | 86 | 81.271 | ENSPANG00000034816 | - | 92 | 81.271 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 88.491 | ENSPANG00000029674 | - | 100 | 88.491 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 90.476 | ENSPANG00000035429 | - | 100 | 90.476 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 88.491 | ENSPANG00000031953 | - | 100 | 88.491 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 87.547 | ENSPANG00000035655 | - | 100 | 87.547 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 89 | 87.764 | ENSPANG00000032780 | - | 100 | 87.764 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 87.547 | ENSPANG00000032302 | - | 100 | 87.547 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 87.170 | ENSPANG00000029660 | - | 100 | 87.170 | Papio_anubis |
| ENSG00000223443 | USP17L2 | 100 | 85.849 | ENSRROG00000032242 | - | 95 | 85.849 | Rhinopithecus_roxellana |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004197 | cysteine-type endopeptidase activity | 21873635. | IBA | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 21873635. | IBA | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 20228808.21239494. | IDA | Function |
| GO:0004843 | thiol-dependent ubiquitin-specific protease activity | 19188362.20368735. | IMP | Function |
| GO:0005515 | protein binding | 21239494. | IPI | Function |
| GO:0005634 | nucleus | 21239494. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | 19188362. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006511 | ubiquitin-dependent protein catabolic process | - | IEA | Process |
| GO:0006915 | apoptotic process | - | IEA | Process |
| GO:0007093 | mitotic cell cycle checkpoint | 20228808. | IMP | Process |
| GO:0010955 | negative regulation of protein processing | 19188362. | IMP | Process |
| GO:0016579 | protein deubiquitination | 21873635. | IBA | Process |
| GO:0016579 | protein deubiquitination | 20228808. | IDA | Process |
| GO:0016579 | protein deubiquitination | - | TAS | Process |
| GO:0030334 | regulation of cell migration | 21448158. | IMP | Process |
| GO:0031064 | negative regulation of histone deacetylation | 21239494. | IMP | Process |
| GO:0034260 | negative regulation of GTPase activity | 19188362. | IDA | Process |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | 20228808. | IMP | Function |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | - | TAS | Function |
| GO:0042127 | regulation of cell proliferation | 19188362. | IMP | Process |
| GO:0042981 | regulation of apoptotic process | 21873635. | IBA | Process |
| GO:0042981 | regulation of apoptotic process | 21239494. | IMP | Process |
| GO:0043547 | positive regulation of GTPase activity | 21448158. | IMP | Process |
| GO:0050691 | regulation of defense response to virus by host | 20368735. | IMP | Process |
| GO:0070536 | protein K63-linked deubiquitination | 21239494. | IDA | Process |
| GO:0070536 | protein K63-linked deubiquitination | 19188362.20368735. | IMP | Process |
| GO:0071108 | protein K48-linked deubiquitination | 19188362.20368735. | IMP | Process |
| GO:0071586 | CAAX-box protein processing | 19188362. | IMP | Process |
| GO:0090315 | negative regulation of protein targeting to membrane | 19188362. | IMP | Process |
| GO:1900027 | regulation of ruffle assembly | 21448158. | IMP | Process |
| GO:1900245 | positive regulation of MDA-5 signaling pathway | 20368735. | IMP | Process |
| GO:1900246 | positive regulation of RIG-I signaling pathway | 20368735. | IMP | Process |