
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| COAD | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| READ | ||
| LAML | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000419901 | Transposase_22 | PF02994.14 | 2.3e-24 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000498273 | L1TD1-201 | 3849 | - | ENSP00000419901 | 865 (aa) | - | Q5T7N2 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000240563 | Bone Density | 9E-6 | 28181694 |
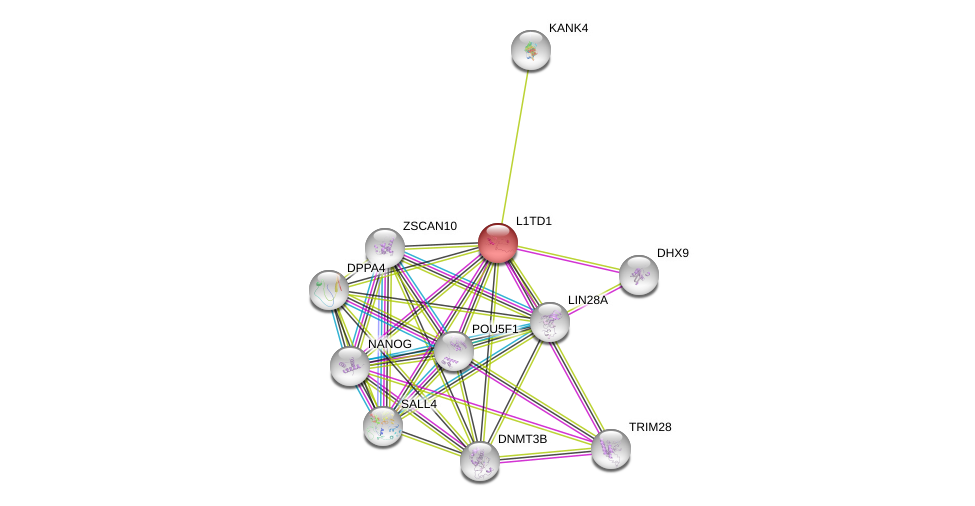
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000240563 | L1TD1 | 99 | 62.572 | ENSAMEG00000014821 | L1TD1 | 99 | 62.442 | Ailuropoda_melanoleuca |
| ENSG00000240563 | L1TD1 | 78 | 85.921 | ENSCJAG00000038509 | L1TD1 | 95 | 87.361 | Callithrix_jacchus |
| ENSG00000240563 | L1TD1 | 54 | 51.200 | ENSCAFG00020012951 | L1TD1 | 58 | 51.005 | Canis_lupus_dingo |
| ENSG00000240563 | L1TD1 | 100 | 87.313 | ENSCATG00000038295 | L1TD1 | 99 | 89.286 | Cercocebus_atys |
| ENSG00000240563 | L1TD1 | 100 | 86.736 | ENSCSAG00000001337 | L1TD1 | 99 | 87.185 | Chlorocebus_sabaeus |
| ENSG00000240563 | L1TD1 | 100 | 88.249 | ENSCANG00000042261 | L1TD1 | 99 | 88.594 | Colobus_angolensis_palliatus |
| ENSG00000240563 | L1TD1 | 99 | 47.680 | ENSCGRG00001024238 | L1td1 | 99 | 47.197 | Cricetulus_griseus_chok1gshd |
| ENSG00000240563 | L1TD1 | 99 | 47.680 | ENSCGRG00000011853 | L1td1 | 99 | 47.197 | Cricetulus_griseus_crigri |
| ENSG00000240563 | L1TD1 | 99 | 54.943 | ENSDNOG00000039645 | - | 100 | 54.891 | Dasypus_novemcinctus |
| ENSG00000240563 | L1TD1 | 99 | 51.185 | ENSDNOG00000040097 | - | 100 | 51.659 | Dasypus_novemcinctus |
| ENSG00000240563 | L1TD1 | 99 | 58.392 | ENSEASG00005008811 | L1TD1 | 63 | 65.426 | Equus_asinus_asinus |
| ENSG00000240563 | L1TD1 | 50 | 37.500 | ENSECAG00000028123 | - | 65 | 40.249 | Equus_caballus |
| ENSG00000240563 | L1TD1 | 99 | 55.923 | ENSECAG00000013248 | L1TD1 | 80 | 64.066 | Equus_caballus |
| ENSG00000240563 | L1TD1 | 99 | 61.601 | ENSFCAG00000042658 | L1TD1 | 99 | 60.343 | Felis_catus |
| ENSG00000240563 | L1TD1 | 100 | 97.347 | ENSGGOG00000026365 | L1TD1 | 100 | 97.347 | Gorilla_gorilla |
| ENSG00000240563 | L1TD1 | 100 | 87.082 | ENSMFAG00000031887 | L1TD1 | 99 | 88.825 | Macaca_fascicularis |
| ENSG00000240563 | L1TD1 | 100 | 88.594 | ENSMNEG00000030098 | L1TD1 | 99 | 88.594 | Macaca_nemestrina |
| ENSG00000240563 | L1TD1 | 100 | 89.516 | ENSMLEG00000039743 | L1TD1 | 99 | 89.516 | Mandrillus_leucophaeus |
| ENSG00000240563 | L1TD1 | 63 | 53.950 | ENSMAUG00000020769 | L1td1 | 60 | 56.349 | Mesocricetus_auratus |
| ENSG00000240563 | L1TD1 | 99 | 68.023 | ENSMICG00000045847 | L1TD1 | 99 | 67.049 | Microcebus_murinus |
| ENSG00000240563 | L1TD1 | 51 | 54.299 | ENSMOCG00000007037 | L1td1 | 56 | 57.500 | Microtus_ochrogaster |
| ENSG00000240563 | L1TD1 | 60 | 51.064 | MGP_CAROLIEiJ_G0026185 | L1td1 | 54 | 54.444 | Mus_caroli |
| ENSG00000240563 | L1TD1 | 60 | 51.179 | ENSMUSG00000087166 | L1td1 | 59 | 52.800 | Mus_musculus |
| ENSG00000240563 | L1TD1 | 60 | 49.531 | MGP_PahariEiJ_G0028522 | L1td1 | 54 | 52.394 | Mus_pahari |
| ENSG00000240563 | L1TD1 | 60 | 50.708 | MGP_SPRETEiJ_G0027153 | L1td1 | 59 | 52.800 | Mus_spretus |
| ENSG00000240563 | L1TD1 | 88 | 50.852 | ENSNGAG00000000776 | L1td1 | 94 | 50.526 | Nannospalax_galili |
| ENSG00000240563 | L1TD1 | 100 | 92.857 | ENSNLEG00000014904 | L1TD1 | 99 | 92.742 | Nomascus_leucogenys |
| ENSG00000240563 | L1TD1 | 100 | 97.575 | ENSPPAG00000043606 | L1TD1 | 100 | 97.575 | Pan_paniscus |
| ENSG00000240563 | L1TD1 | 99 | 61.547 | ENSPPRG00000011113 | L1TD1 | 99 | 61.618 | Panthera_pardus |
| ENSG00000240563 | L1TD1 | 100 | 97.806 | ENSPTRG00000034387 | L1TD1 | 100 | 97.806 | Pan_troglodytes |
| ENSG00000240563 | L1TD1 | 100 | 87.082 | ENSPANG00000021627 | L1TD1 | 99 | 87.872 | Papio_anubis |
| ENSG00000240563 | L1TD1 | 99 | 45.244 | ENSPEMG00000008068 | L1td1 | 99 | 46.599 | Peromyscus_maniculatus_bairdii |
| ENSG00000240563 | L1TD1 | 100 | 91.109 | ENSPPYG00000001286 | L1TD1 | 99 | 91.109 | Pongo_abelii |
| ENSG00000240563 | L1TD1 | 99 | 68.894 | ENSPCOG00000005879 | L1TD1 | 99 | 70.298 | Propithecus_coquereli |
| ENSG00000240563 | L1TD1 | 100 | 87.529 | ENSRBIG00000005782 | L1TD1 | 99 | 87.774 | Rhinopithecus_bieti |
| ENSG00000240563 | L1TD1 | 100 | 87.659 | ENSRROG00000042567 | L1TD1 | 99 | 88.005 | Rhinopithecus_roxellana |
| ENSG00000240563 | L1TD1 | 78 | 63.696 | ENSTBEG00000015274 | L1TD1 | 99 | 48.092 | Tupaia_belangeri |
| ENSG00000240563 | L1TD1 | 99 | 62.121 | ENSUAMG00000002306 | L1TD1 | 99 | 60.950 | Ursus_americanus |
| ENSG00000240563 | L1TD1 | 99 | 57.293 | ENSUMAG00000014924 | L1TD1 | 99 | 58.460 | Ursus_maritimus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003727 | single-stranded RNA binding | 21873635. | IBA | Function |
| GO:0032197 | transposition, RNA-mediated | 21873635. | IBA | Process |
| GO:1990904 | ribonucleoprotein complex | 21873635. | IBA | Component |