
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| CESC | |||||||
| LUAD | |||||||
| OV | |||||||
| SARC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| GBM | ||
| CHOL | ||
| LUAD | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000308472 | eIF_4EBP | PF05456.11 | 1.4e-37 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000310331 | EIF4EBP3-201 | 691 | - | ENSP00000308472 | 100 (aa) | - | O60516 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03013 | RNA transport | KEGG |
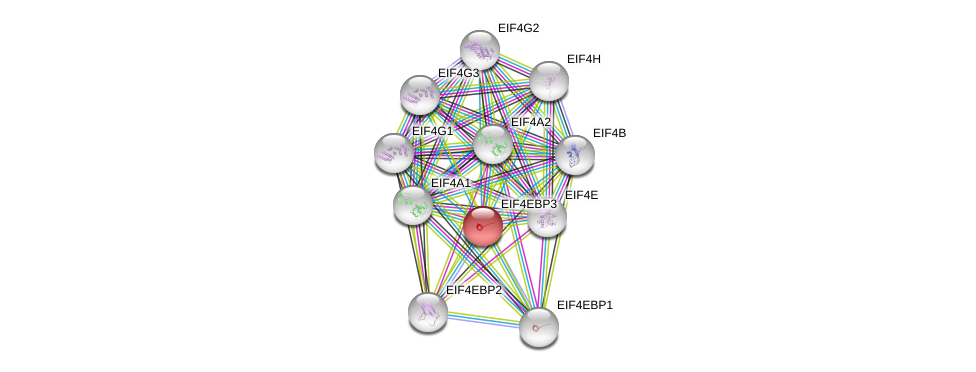
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSAPOG00000010355 | eif4ebp3l | 100 | 65.179 | Acanthochromis_polyacanthus |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSAOCG00000007351 | eif4ebp3l | 100 | 65.179 | Amphiprion_ocellaris |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSAPEG00000010500 | eif4ebp3l | 100 | 65.179 | Amphiprion_percula |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.071 | ENSATEG00000000149 | eif4ebp3l | 100 | 64.286 | Anabas_testudineus |
| ENSG00000243056 | EIF4EBP3 | 86 | 75.000 | ENSACLG00000025145 | eif4ebp3l | 79 | 72.727 | Astatotilapia_calliptera |
| ENSG00000243056 | EIF4EBP3 | 97 | 62.136 | ENSAMXG00000007416 | eif4ebp3 | 94 | 61.165 | Astyanax_mexicanus |
| ENSG00000243056 | EIF4EBP3 | 100 | 61.607 | ENSAMXG00000042158 | eif4ebp3l | 100 | 66.964 | Astyanax_mexicanus |
| ENSG00000243056 | EIF4EBP3 | 76 | 85.714 | ENSBTAG00000052310 | EIF4EBP3 | 74 | 81.818 | Bos_taurus |
| ENSG00000243056 | EIF4EBP3 | 100 | 99.000 | ENSCJAG00000041408 | EIF4EBP3 | 100 | 99.000 | Callithrix_jacchus |
| ENSG00000243056 | EIF4EBP3 | 100 | 98.000 | ENSCAFG00020001947 | EIF4EBP3 | 100 | 98.000 | Canis_lupus_dingo |
| ENSG00000243056 | EIF4EBP3 | 87 | 79.348 | ENSCHIG00000026608 | EIF4EBP3 | 90 | 76.087 | Capra_hircus |
| ENSG00000243056 | EIF4EBP3 | 100 | 88.000 | ENSTSYG00000028184 | EIF4EBP3 | 100 | 88.000 | Carlito_syrichta |
| ENSG00000243056 | EIF4EBP3 | 100 | 90.099 | ENSCPOG00000033979 | EIF4EBP3 | 100 | 86.139 | Cavia_porcellus |
| ENSG00000243056 | EIF4EBP3 | 100 | 88.119 | ENSCLAG00000003391 | EIF4EBP3 | 100 | 84.158 | Chinchilla_lanigera |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.000 | ENSCSAG00000012830 | EIF4EBP3 | 100 | 97.000 | Chlorocebus_sabaeus |
| ENSG00000243056 | EIF4EBP3 | 99 | 80.198 | ENSCPBG00000007581 | EIF4EBP3 | 96 | 76.238 | Chrysemys_picta_bellii |
| ENSG00000243056 | EIF4EBP3 | 99 | 98.990 | ENSCANG00000036245 | EIF4EBP3 | 100 | 98.990 | Colobus_angolensis_palliatus |
| ENSG00000243056 | EIF4EBP3 | 100 | 86.139 | ENSCGRG00001014184 | Eif4ebp3 | 100 | 82.178 | Cricetulus_griseus_chok1gshd |
| ENSG00000243056 | EIF4EBP3 | 100 | 86.139 | ENSCGRG00000006941 | Eif4ebp3 | 100 | 82.178 | Cricetulus_griseus_crigri |
| ENSG00000243056 | EIF4EBP3 | 100 | 63.393 | ENSCSEG00000018681 | eif4ebp3l | 100 | 63.393 | Cynoglossus_semilaevis |
| ENSG00000243056 | EIF4EBP3 | 95 | 66.667 | ENSCVAG00000021288 | eif4ebp3l | 92 | 64.762 | Cyprinodon_variegatus |
| ENSG00000243056 | EIF4EBP3 | 97 | 57.692 | ENSDARG00000054916 | eif4ebp3 | 94 | 58.252 | Danio_rerio |
| ENSG00000243056 | EIF4EBP3 | 100 | 63.393 | ENSDARG00000041607 | eif4ebp3l | 100 | 65.179 | Danio_rerio |
| ENSG00000243056 | EIF4EBP3 | 100 | 91.089 | ENSDORG00000024746 | Eif4ebp3 | 100 | 87.129 | Dipodomys_ordii |
| ENSG00000243056 | EIF4EBP3 | 100 | 94.059 | ENSEASG00005017482 | EIF4EBP3 | 100 | 90.099 | Equus_asinus_asinus |
| ENSG00000243056 | EIF4EBP3 | 100 | 93.069 | ENSECAG00000034514 | EIF4EBP3 | 100 | 89.109 | Equus_caballus |
| ENSG00000243056 | EIF4EBP3 | 95 | 67.647 | ENSELUG00000006596 | eif4ebp3l | 92 | 71.569 | Esox_lucius |
| ENSG00000243056 | EIF4EBP3 | 95 | 64.356 | ENSELUG00000021245 | eif4ebp3 | 94 | 63.366 | Esox_lucius |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.030 | ENSFCAG00000044383 | EIF4EBP3 | 100 | 97.030 | Felis_catus |
| ENSG00000243056 | EIF4EBP3 | 100 | 93.069 | ENSFDAG00000013042 | EIF4EBP3 | 100 | 91.089 | Fukomys_damarensis |
| ENSG00000243056 | EIF4EBP3 | 100 | 64.035 | ENSFHEG00000001957 | eif4ebp3l | 100 | 62.281 | Fundulus_heteroclitus |
| ENSG00000243056 | EIF4EBP3 | 100 | 68.182 | ENSGMOG00000003122 | eif4ebp3l | 96 | 65.421 | Gadus_morhua |
| ENSG00000243056 | EIF4EBP3 | 67 | 70.588 | ENSGALG00000048830 | EIF4EBP3 | 55 | 64.706 | Gallus_gallus |
| ENSG00000243056 | EIF4EBP3 | 82 | 67.021 | ENSGAFG00000002815 | eif4ebp3l | 75 | 74.667 | Gambusia_affinis |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSGACG00000018498 | eif4ebp3l | 98 | 65.179 | Gasterosteus_aculeatus |
| ENSG00000243056 | EIF4EBP3 | 86 | 75.000 | ENSHBUG00000003021 | eif4ebp3l | 79 | 72.727 | Haplochromis_burtoni |
| ENSG00000243056 | EIF4EBP3 | 100 | 95.050 | ENSHGLG00000008728 | EIF4EBP3 | 100 | 91.089 | Heterocephalus_glaber_female |
| ENSG00000243056 | EIF4EBP3 | 100 | 95.050 | ENSHGLG00100006719 | EIF4EBP3 | 100 | 91.089 | Heterocephalus_glaber_male |
| ENSG00000243056 | EIF4EBP3 | 100 | 65.455 | ENSHCOG00000003055 | eif4ebp3l | 100 | 65.455 | Hippocampus_comes |
| ENSG00000243056 | EIF4EBP3 | 100 | 63.393 | ENSIPUG00000018246 | 4eb3l | 100 | 70.536 | Ictalurus_punctatus |
| ENSG00000243056 | EIF4EBP3 | 98 | 61.321 | ENSIPUG00000020570 | eif4ebp3l | 94 | 60.952 | Ictalurus_punctatus |
| ENSG00000243056 | EIF4EBP3 | 100 | 92.079 | ENSSTOG00000020350 | EIF4EBP3 | 100 | 88.119 | Ictidomys_tridecemlineatus |
| ENSG00000243056 | EIF4EBP3 | 100 | 92.079 | ENSJJAG00000016625 | Eif4ebp3 | 100 | 84.000 | Jaculus_jaculus |
| ENSG00000243056 | EIF4EBP3 | 100 | 67.257 | ENSKMAG00000016697 | eif4ebp3l | 100 | 66.372 | Kryptolebias_marmoratus |
| ENSG00000243056 | EIF4EBP3 | 95 | 67.961 | ENSLBEG00000014223 | eif4ebp3l | 92 | 66.990 | Labrus_bergylta |
| ENSG00000243056 | EIF4EBP3 | 100 | 72.897 | ENSLACG00000016472 | EIF4EBP3 | 96 | 71.963 | Latimeria_chalumnae |
| ENSG00000243056 | EIF4EBP3 | 80 | 74.419 | ENSLOCG00000012500 | eif4ebp3l | 89 | 74.074 | Lepisosteus_oculatus |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.071 | ENSMAMG00000013532 | eif4ebp3l | 100 | 64.286 | Mastacembelus_armatus |
| ENSG00000243056 | EIF4EBP3 | 86 | 75.000 | ENSMZEG00005007636 | eif4ebp3l | 79 | 72.727 | Maylandia_zebra |
| ENSG00000243056 | EIF4EBP3 | 100 | 85.149 | ENSMAUG00000015124 | Eif4ebp3 | 100 | 81.188 | Mesocricetus_auratus |
| ENSG00000243056 | EIF4EBP3 | 100 | 80.198 | ENSMOCG00000021408 | Eif4ebp3 | 100 | 74.000 | Microtus_ochrogaster |
| ENSG00000243056 | EIF4EBP3 | 99 | 66.355 | ENSMMOG00000009467 | eif4ebp3l | 96 | 64.486 | Mola_mola |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.071 | ENSMALG00000006261 | eif4ebp3l | 100 | 65.179 | Monopterus_albus |
| ENSG00000243056 | EIF4EBP3 | 100 | 87.129 | MGP_CAROLIEiJ_G0022128 | Eif4ebp3 | 100 | 83.168 | Mus_caroli |
| ENSG00000243056 | EIF4EBP3 | 100 | 86.139 | ENSMUSG00000090264 | Eif4ebp3 | 100 | 82.178 | Mus_musculus |
| ENSG00000243056 | EIF4EBP3 | 100 | 87.129 | MGP_PahariEiJ_G0018862 | Eif4ebp3 | 100 | 83.168 | Mus_pahari |
| ENSG00000243056 | EIF4EBP3 | 100 | 87.129 | MGP_SPRETEiJ_G0023040 | Eif4ebp3 | 100 | 83.168 | Mus_spretus |
| ENSG00000243056 | EIF4EBP3 | 100 | 83.168 | ENSNGAG00000016353 | Eif4ebp3 | 100 | 85.149 | Nannospalax_galili |
| ENSG00000243056 | EIF4EBP3 | 100 | 92.079 | ENSODEG00000004026 | EIF4EBP3 | 100 | 88.119 | Octodon_degus |
| ENSG00000243056 | EIF4EBP3 | 86 | 75.000 | ENSONIG00000017571 | eif4ebp3l | 79 | 72.727 | Oreochromis_niloticus |
| ENSG00000243056 | EIF4EBP3 | 73 | 86.486 | ENSOANG00000001494 | EIF4EBP3 | 100 | 81.081 | Ornithorhynchus_anatinus |
| ENSG00000243056 | EIF4EBP3 | 97 | 68.932 | ENSORLG00000024341 | eif4ebp3l | 94 | 66.019 | Oryzias_latipes |
| ENSG00000243056 | EIF4EBP3 | 97 | 68.932 | ENSORLG00020004753 | eif4ebp3l | 94 | 66.019 | Oryzias_latipes_hni |
| ENSG00000243056 | EIF4EBP3 | 97 | 68.932 | ENSORLG00015008086 | eif4ebp3l | 94 | 66.019 | Oryzias_latipes_hsok |
| ENSG00000243056 | EIF4EBP3 | 97 | 68.932 | ENSOMEG00000013335 | eif4ebp3l | 94 | 67.961 | Oryzias_melastigma |
| ENSG00000243056 | EIF4EBP3 | 100 | 96.040 | ENSPPRG00000023846 | EIF4EBP3 | 100 | 96.040 | Panthera_pardus |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.030 | ENSPTIG00000010875 | EIF4EBP3 | 100 | 97.030 | Panthera_tigris_altaica |
| ENSG00000243056 | EIF4EBP3 | 70 | 72.368 | ENSPKIG00000005779 | eif4ebp3 | 60 | 68.675 | Paramormyrops_kingsleyae |
| ENSG00000243056 | EIF4EBP3 | 86 | 77.011 | ENSPMGG00000000673 | eif4ebp3l | 75 | 79.310 | Periophthalmus_magnuspinnatus |
| ENSG00000243056 | EIF4EBP3 | 100 | 79.000 | ENSPCIG00000017473 | EIF4EBP3 | 100 | 79.000 | Phascolarctos_cinereus |
| ENSG00000243056 | EIF4EBP3 | 100 | 64.912 | ENSPFOG00000007698 | eif4ebp3l | 100 | 63.158 | Poecilia_formosa |
| ENSG00000243056 | EIF4EBP3 | 82 | 67.021 | ENSPLAG00000002047 | eif4ebp3l | 74 | 71.951 | Poecilia_latipinna |
| ENSG00000243056 | EIF4EBP3 | 100 | 64.912 | ENSPMEG00000008305 | eif4ebp3l | 100 | 63.158 | Poecilia_mexicana |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.030 | ENSPCOG00000014289 | EIF4EBP3 | 100 | 97.030 | Propithecus_coquereli |
| ENSG00000243056 | EIF4EBP3 | 99 | 63.810 | ENSPNAG00000007774 | eif4ebp3l | 90 | 66.000 | Pygocentrus_nattereri |
| ENSG00000243056 | EIF4EBP3 | 98 | 59.615 | ENSPNAG00000011458 | eif4ebp3 | 95 | 59.048 | Pygocentrus_nattereri |
| ENSG00000243056 | EIF4EBP3 | 87 | 68.539 | ENSSFOG00015016325 | eif4ebp3l | 94 | 76.404 | Scleropages_formosus |
| ENSG00000243056 | EIF4EBP3 | 97 | 67.619 | ENSSMAG00000010258 | eif4ebp3l | 95 | 66.667 | Scophthalmus_maximus |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSSDUG00000011012 | eif4ebp3l | 100 | 65.179 | Seriola_dumerili |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSSLDG00000002841 | eif4ebp3l | 100 | 65.179 | Seriola_lalandi_dorsalis |
| ENSG00000243056 | EIF4EBP3 | 100 | 66.964 | ENSSPAG00000015188 | eif4ebp3l | 100 | 65.179 | Stegastes_partitus |
| ENSG00000243056 | EIF4EBP3 | 100 | 93.069 | ENSSSCG00000020835 | EIF4EBP3 | 100 | 92.079 | Sus_scrofa |
| ENSG00000243056 | EIF4EBP3 | 95 | 67.961 | ENSTRUG00000002444 | eif4ebp3l | 92 | 66.019 | Takifugu_rubripes |
| ENSG00000243056 | EIF4EBP3 | 95 | 66.990 | ENSTNIG00000015789 | eif4ebp3l | 91 | 66.019 | Tetraodon_nigroviridis |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.030 | ENSUAMG00000005352 | EIF4EBP3 | 100 | 97.030 | Ursus_americanus |
| ENSG00000243056 | EIF4EBP3 | 100 | 97.030 | ENSUMAG00000013279 | EIF4EBP3 | 100 | 97.030 | Ursus_maritimus |
| ENSG00000243056 | EIF4EBP3 | 100 | 96.040 | ENSVVUG00000012362 | - | 100 | 96.040 | Vulpes_vulpes |
| ENSG00000243056 | EIF4EBP3 | 95 | 64.583 | ENSVVUG00000015501 | - | 62 | 81.250 | Vulpes_vulpes |
| ENSG00000243056 | EIF4EBP3 | 100 | 64.912 | ENSXCOG00000011168 | eif4ebp3l | 100 | 63.158 | Xiphophorus_couchianus |
| ENSG00000243056 | EIF4EBP3 | 100 | 64.912 | ENSXMAG00000029844 | eif4ebp3l | 100 | 63.158 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 15153109.16271312.22684010.25416956. | IPI | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0008190 | eukaryotic initiation factor 4E binding | 21873635. | IBA | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016281 | eukaryotic translation initiation factor 4F complex | 9593750. | NAS | Component |
| GO:0030371 | translation repressor activity | 9593750. | NAS | Function |
| GO:0045947 | negative regulation of translational initiation | 21873635. | IBA | Process |
| GO:0045947 | negative regulation of translational initiation | 9593750. | NAS | Process |