
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27609572 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| DLBC | |||
| PAAD | |||
| READ | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| HNSC | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| UCEC | ||
| LGG | ||
| THCA | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000432695 | Methyltransf_31 | PF13847.6 | 3e-09 | 1 | 1 |
| ENSP00000432695 | Methyltransf_8 | PF05148.15 | 3.7e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000532376 | TRMT9B-209 | 668 | - | ENSP00000431717 | 54 (aa) | - | E9PK20 |
| ENST00000400069 | TRMT9B-201 | 1982 | - | ENSP00000468715 | 60 (aa) | - | Q9P272 |
| ENST00000525249 | TRMT9B-204 | 553 | - | - | - (aa) | - | - |
| ENST00000529978 | TRMT9B-208 | 6003 | - | - | - (aa) | - | - |
| ENST00000447063 | TRMT9B-202 | 983 | - | ENSP00000443288 | 196 (aa) | - | F6XBY7 |
| ENST00000524591 | TRMT9B-203 | 9587 | XM_005273584 | ENSP00000432695 | 454 (aa) | XP_005273641 | Q9P272 |
| ENST00000529706 | TRMT9B-207 | 2758 | - | - | - (aa) | - | - |
| ENST00000528753 | TRMT9B-206 | 2392 | - | ENSP00000466330 | 60 (aa) | - | Q9P272 |
| ENST00000528335 | TRMT9B-205 | 579 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000250305 | Arteries | 1.9560000E-005 | - |
| ENSG00000250305 | Platelet Function Tests | 1.4169400E-005 | - |
| ENSG00000250305 | Child Development Disorders, Pervasive | 6.1462000E-006 | - |
| ENSG00000250305 | Child Development Disorders, Pervasive | 3.7093000E-006 | - |
| ENSG00000250305 | Child Development Disorders, Pervasive | 1.5734000E-005 | - |
| ENSG00000250305 | Child Development Disorders, Pervasive | 2.0084000E-005 | - |
| ENSG00000250305 | S-(3-hydroxypropyl)cysteine N-acetate | 3E-6 | 26053186 |
| ENSG00000250305 | Smoking | 3E-6 | 26053186 |
| ENSG00000250305 | Diabetes Mellitus, Type 2 | 4E-6 | 26818947 |
| ENSG00000250305 | Bronchodilator Agents | 2E-6 | 26634245 |
| ENSG00000250305 | Lung Volume Measurements | 2E-6 | 26634245 |
| ENSG00000250305 | Migraine Disorders | 9E-6 | 23793025 |
| ENSG00000250305 | Bronchodilator Agents | 2E-6 | 26634245 |
| ENSG00000250305 | Lung Volume Measurements | 2E-6 | 26634245 |
| ENSG00000250305 | Stress Disorders, Post-Traumatic | 5E-6 | 24677629 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000250305 | rs2946504 | 8 | 12954071 | T | Type 2 diabetes | 26818947 | [1.04-1.09] | 1.06 | EFO_0001360 |
| ENSG00000250305 | rs2946505 | 8 | 12953643 | ? | Migraine | 23793025 | [1.03-1.09] | 1.06 | EFO_0003821 |
| ENSG00000250305 | rs2460905 | 8 | 12977091 | ? | Post-traumatic stress disorder | 24677629 | [0.79-1.97] unit increase | 1.38 | EFO_0001358 |
| ENSG00000250305 | rs2466254 | 8 | 12976853 | ? | Pneumonia | 28928442 | [0.029-0.073] unit increase | 0.0507 | EFO_0008410 |
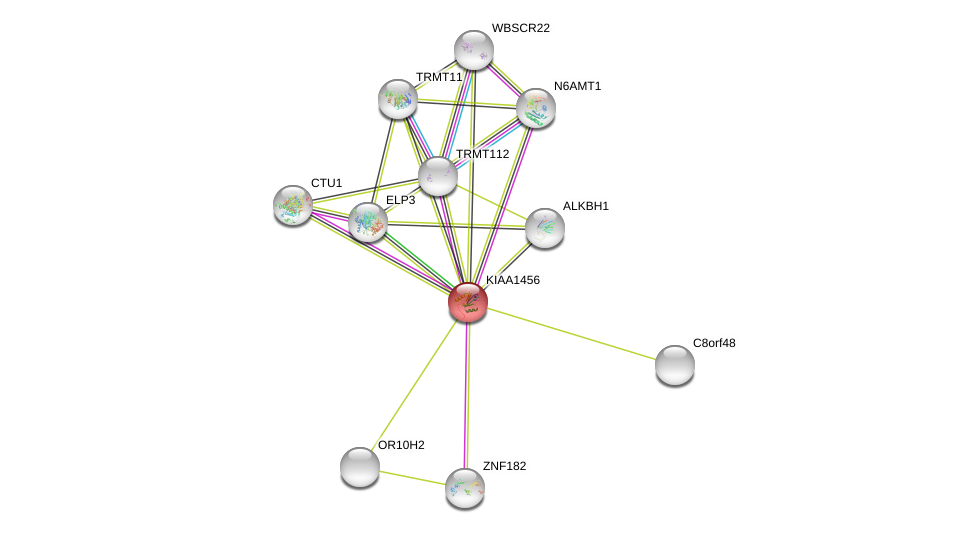
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSAMEG00000007166 | TRMT9B | 99 | 81.111 | Ailuropoda_melanoleuca |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | ENSAPLG00000009756 | TRMT9B | 99 | 60.435 | Anas_platyrhynchos |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSANAG00000021322 | TRMT9B | 100 | 93.172 | Aotus_nancymaae |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSBTAG00000008950 | TRMT9B | 100 | 78.855 | Bos_taurus |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSCJAG00000012066 | TRMT9B | 100 | 92.511 | Callithrix_jacchus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSCAFG00000006759 | TRMT9B | 100 | 82.159 | Canis_familiaris |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSCHIG00000025183 | TRMT9B | 100 | 79.515 | Capra_hircus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSTSYG00000029308 | TRMT9B | 100 | 85.683 | Carlito_syrichta |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSCPOG00000032099 | TRMT9B | 99 | 76.222 | Cavia_porcellus |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSCCAG00000032783 | TRMT9B | 100 | 92.731 | Cebus_capucinus |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSCATG00000037514 | TRMT9B | 100 | 94.714 | Cercocebus_atys |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSCLAG00000000634 | TRMT9B | 99 | 79.333 | Chinchilla_lanigera |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSCSAG00000016833 | TRMT9B | 100 | 94.273 | Chlorocebus_sabaeus |
| ENSG00000250305 | TRMT9B | 100 | 87.037 | ENSCPBG00000025129 | TRMT9B | 97 | 62.472 | Chrysemys_picta_bellii |
| ENSG00000250305 | TRMT9B | 89 | 93.781 | ENSCANG00000025271 | TRMT9B | 100 | 93.781 | Colobus_angolensis_palliatus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSCGRG00001019764 | Trmt9b | 99 | 72.907 | Cricetulus_griseus_chok1gshd |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSCGRG00000012855 | Trmt9b | 99 | 72.907 | Cricetulus_griseus_crigri |
| ENSG00000250305 | TRMT9B | 100 | 85.185 | ENSDNOG00000000297 | TRMT9B | 100 | 73.789 | Dasypus_novemcinctus |
| ENSG00000250305 | TRMT9B | 100 | 90.741 | ENSDORG00000013664 | - | 100 | 88.073 | Dipodomys_ordii |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSEASG00005017014 | TRMT9B | 100 | 83.297 | Equus_asinus_asinus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSECAG00000013811 | TRMT9B | 100 | 83.700 | Equus_caballus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSFCAG00000041381 | TRMT9B | 100 | 81.938 | Felis_catus |
| ENSG00000250305 | TRMT9B | 100 | 81.481 | ENSFALG00000003704 | TRMT9B | 98 | 59.695 | Ficedula_albicollis |
| ENSG00000250305 | TRMT9B | 100 | 87.037 | ENSFDAG00000020217 | TRMT9B | 99 | 78.667 | Fukomys_damarensis |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | ENSGALG00000042726 | TRMT9B | 99 | 59.737 | Gallus_gallus |
| ENSG00000250305 | TRMT9B | 100 | 87.037 | ENSGAGG00000006042 | TRMT9B | 99 | 61.062 | Gopherus_agassizii |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSGGOG00000037983 | TRMT9B | 100 | 98.238 | Gorilla_gorilla |
| ENSG00000250305 | TRMT9B | 100 | 88.889 | ENSHGLG00000004927 | TRMT9B | 99 | 81.778 | Heterocephalus_glaber_female |
| ENSG00000250305 | TRMT9B | 100 | 90.741 | ENSHGLG00100001579 | TRMT9B | 99 | 82.000 | Heterocephalus_glaber_male |
| ENSG00000250305 | TRMT9B | 100 | 79.630 | ENSLACG00000009737 | TRMT9B | 99 | 54.013 | Latimeria_chalumnae |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSMFAG00000043419 | TRMT9B | 100 | 94.273 | Macaca_fascicularis |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSMMUG00000043856 | TRMT9B | 100 | 94.273 | Macaca_mulatta |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSMNEG00000037430 | TRMT9B | 100 | 94.273 | Macaca_nemestrina |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSMLEG00000037379 | TRMT9B | 100 | 94.273 | Mandrillus_leucophaeus |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | ENSMGAG00000011654 | TRMT9B | 99 | 60.613 | Meleagris_gallopavo |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSMAUG00000017514 | Trmt9b | 99 | 72.667 | Mesocricetus_auratus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSMICG00000012942 | TRMT9B | 100 | 84.141 | Microcebus_murinus |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSMOCG00000013997 | Trmt9b | 99 | 70.222 | Microtus_ochrogaster |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | MGP_CAROLIEiJ_G0030985 | Trmt9b | 100 | 74.066 | Mus_caroli |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | ENSMUSG00000039620 | Trmt9b | 100 | 73.568 | Mus_musculus |
| ENSG00000250305 | TRMT9B | 100 | 88.889 | MGP_PahariEiJ_G0021453 | Trmt9b | 100 | 73.789 | Mus_pahari |
| ENSG00000250305 | TRMT9B | 100 | 83.333 | MGP_SPRETEiJ_G0032101 | Trmt9b | 100 | 73.348 | Mus_spretus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSMPUG00000011528 | TRMT9B | 100 | 79.075 | Mustela_putorius_furo |
| ENSG00000250305 | TRMT9B | 100 | 88.889 | ENSMLUG00000013029 | TRMT9B | 100 | 81.278 | Myotis_lucifugus |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSNLEG00000031644 | TRMT9B | 100 | 95.154 | Nomascus_leucogenys |
| ENSG00000250305 | TRMT9B | 100 | 90.741 | ENSODEG00000004422 | TRMT9B | 99 | 80.222 | Octodon_degus |
| ENSG00000250305 | TRMT9B | 100 | 71.186 | ENSOANG00000002169 | - | 79 | 57.034 | Ornithorhynchus_anatinus |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSOCUG00000024898 | TRMT9B | 100 | 77.074 | Oryctolagus_cuniculus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSOGAG00000012709 | TRMT9B | 100 | 82.159 | Otolemur_garnettii |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSOARG00000009832 | TRMT9B | 99 | 79.379 | Ovis_aries |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSPPAG00000030027 | TRMT9B | 100 | 97.577 | Pan_paniscus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSPPRG00000012143 | TRMT9B | 100 | 82.379 | Panthera_pardus |
| ENSG00000250305 | TRMT9B | 100 | 94.444 | ENSPTIG00000001444 | TRMT9B | 100 | 82.599 | Panthera_tigris_altaica |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSPTRG00000041473 | TRMT9B | 100 | 97.797 | Pan_troglodytes |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSPANG00000008717 | TRMT9B | 100 | 94.273 | Papio_anubis |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSPEMG00000021287 | Trmt9b | 87 | 91.892 | Peromyscus_maniculatus_bairdii |
| ENSG00000250305 | TRMT9B | 100 | 98.148 | ENSPCOG00000020130 | TRMT9B | 100 | 85.055 | Propithecus_coquereli |
| ENSG00000250305 | TRMT9B | 100 | 90.741 | ENSRNOG00000011253 | Trmt9b | 100 | 74.229 | Rattus_norvegicus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSRBIG00000040588 | TRMT9B | 100 | 93.612 | Rhinopithecus_bieti |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSRROG00000005726 | TRMT9B | 100 | 93.392 | Rhinopithecus_roxellana |
| ENSG00000250305 | TRMT9B | 100 | 100.000 | ENSSBOG00000029215 | TRMT9B | 100 | 92.291 | Saimiri_boliviensis_boliviensis |
| ENSG00000250305 | TRMT9B | 93 | 86.000 | ENSSHAG00000001652 | TRMT9B | 98 | 51.663 | Sarcophilus_harrisii |
| ENSG00000250305 | TRMT9B | 100 | 92.593 | ENSSSCG00000006969 | TRMT9B | 100 | 79.956 | Sus_scrofa |
| ENSG00000250305 | TRMT9B | 100 | 81.481 | ENSTGUG00000007372 | TRMT9B | 99 | 58.913 | Taeniopygia_guttata |
| ENSG00000250305 | TRMT9B | 100 | 90.741 | ENSUMAG00000020549 | TRMT9B | 100 | 79.956 | Ursus_maritimus |
| ENSG00000250305 | TRMT9B | 100 | 96.296 | ENSVVUG00000019651 | TRMT9B | 100 | 81.718 | Vulpes_vulpes |
| ENSG00000250305 | TRMT9B | 100 | 79.630 | ENSXETG00000005622 | kiaa1456l | 99 | 50.873 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000049 | tRNA binding | 21873635. | IBA | Function |
| GO:0002098 | tRNA wobble uridine modification | 21873635. | IBA | Process |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | - | TAS | Component |
| GO:0006400 | tRNA modification | - | TAS | Process |
| GO:0008175 | tRNA methyltransferase activity | 23381944. | EXP | Function |
| GO:0016300 | tRNA (uracil) methyltransferase activity | 21873635. | IBA | Function |
| GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 21873635. | IBA | Function |
| GO:0030488 | tRNA methylation | - | IEA | Process |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |