
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000586407 | LIG3-208 | 815 | - | - | - (aa) | - | - |
| ENST00000592690 | LIG3-215 | 722 | - | ENSP00000467063 | 207 (aa) | - | K7ENR9 |
| ENST00000262327 | LIG3-201 | 3055 | XM_006721896 | ENSP00000262327 | 949 (aa) | XP_006721959 | P49916 |
| ENST00000378526 | LIG3-202 | 8400 | XM_005257970 | ENSP00000367787 | 1009 (aa) | XP_005258027 | P49916 |
| ENST00000586119 | LIG3-207 | 565 | - | - | - (aa) | - | - |
| ENST00000585370 | LIG3-203 | 235 | - | ENSP00000465288 | 36 (aa) | - | K7EJR4 |
| ENST00000592244 | LIG3-214 | 440 | - | - | - (aa) | - | - |
| ENST00000586435 | LIG3-209 | 588 | - | - | - (aa) | - | - |
| ENST00000585740 | LIG3-204 | 887 | - | ENSP00000467757 | 185 (aa) | - | K7EQB6 |
| ENST00000593099 | LIG3-216 | 4664 | - | - | - (aa) | - | - |
| ENST00000588713 | LIG3-211 | 467 | - | - | - (aa) | - | - |
| ENST00000590181 | LIG3-212 | 1156 | - | - | - (aa) | - | - |
| ENST00000586058 | LIG3-206 | 581 | - | - | - (aa) | - | - |
| ENST00000590630 | LIG3-213 | 904 | - | - | - (aa) | - | - |
| ENST00000588109 | LIG3-210 | 592 | - | ENSP00000466278 | 38 (aa) | - | K7ELY6 |
| ENST00000585941 | LIG3-205 | 1699 | - | ENSP00000468479 | 545 (aa) | - | K7ERZ5 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03410 | Base excision repair | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000005156 | rs1052536 | 17 | 35004556 | C | QT interval | 24952745 | [0.78-1.18] unit increase | 0.98 | EFO_0004682 |
| ENSG00000005156 | rs2074518 | 17 | 34997363 | T | QT interval | 19305408 | [0.71-1.39] msec decrease | 1.05 | EFO_0004682 |
| ENSG00000005156 | rs145583519 | 17 | 34981227 | G | Macrophage inflammatory protein 1b levels | 27989323 | [0.29-0.52] SD units increase | 0.4044 | EFO_0008219 |
| ENSG00000005156 | rs3135977 | 17 | 34990229 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 | ||
| ENSG00000005156 | rs3135967 | 17 | 34986710 | A | Pulse pressure | 30224653 | [0.079-0.147] unit increase | 0.1128 | EFO_0005763 |
| ENSG00000005156 | rs2074518 | 17 | 34997363 | T | QT interval | 30679814 | [0.97-1.69] unit decrease | 1.33 | EFO_0004682 |
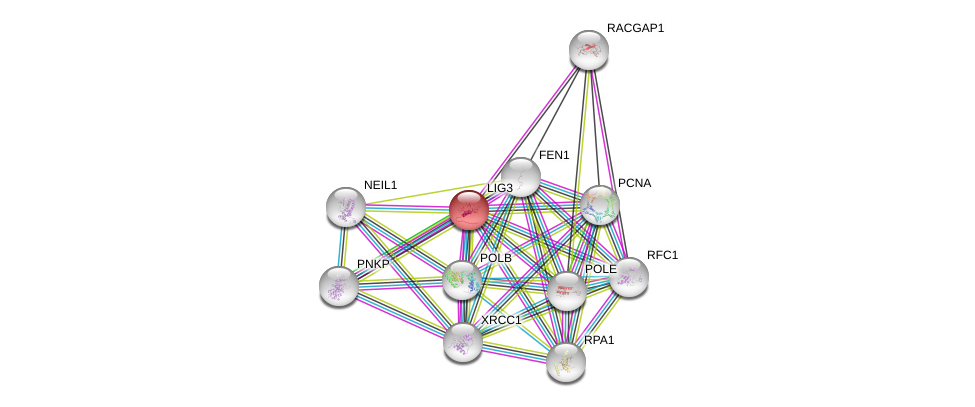
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000724 | double-strand break repair via homologous recombination | - | TAS | Process |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003909 | DNA ligase activity | 9809069. | IDA | Function |
| GO:0003909 | DNA ligase activity | - | TAS | Function |
| GO:0003910 | DNA ligase (ATP) activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 15744309.24407287.25609649. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0006273 | lagging strand elongation | 21873635. | IBA | Process |
| GO:0006283 | transcription-coupled nucleotide-excision repair | - | TAS | Process |
| GO:0006288 | base-excision repair, DNA ligation | 21873635. | IBA | Process |
| GO:0006288 | base-excision repair, DNA ligation | 19589734. | IDA | Process |
| GO:0006288 | base-excision repair, DNA ligation | - | TAS | Process |
| GO:0006297 | nucleotide-excision repair, DNA gap filling | - | TAS | Process |
| GO:0006302 | double-strand break repair | 19589734. | IDA | Process |
| GO:0007049 | cell cycle | - | IEA | Process |
| GO:0008270 | zinc ion binding | - | IEA | Function |
| GO:0033151 | V(D)J recombination | 9809069. | IDA | Process |
| GO:0043504 | mitochondrial DNA repair | 21873635. | IBA | Process |
| GO:0051301 | cell division | - | IEA | Process |
| GO:0071897 | DNA biosynthetic process | - | IEA | Process |
| GO:0090298 | negative regulation of mitochondrial DNA replication | 21878356. | IMP | Process |
| GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 21873635. | IBA | Process |