
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000007699 | CSD | PF00313.22 | 3.9e-25 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000571127 | YBX2-204 | 791 | - | - | - (aa) | - | - |
| ENST00000571834 | YBX2-207 | 2195 | - | - | - (aa) | - | - |
| ENST00000570627 | YBX2-202 | 653 | - | - | - (aa) | - | - |
| ENST00000571464 | YBX2-205 | 640 | - | ENSP00000459587 | 164 (aa) | - | I3L2D6 |
| ENST00000571485 | YBX2-206 | 4553 | - | - | - (aa) | - | - |
| ENST00000007699 | YBX2-201 | 1590 | XM_017024713 | ENSP00000007699 | 364 (aa) | XP_016880202 | Q9Y2T7 |
| ENST00000570720 | YBX2-203 | 606 | - | - | - (aa) | - | - |
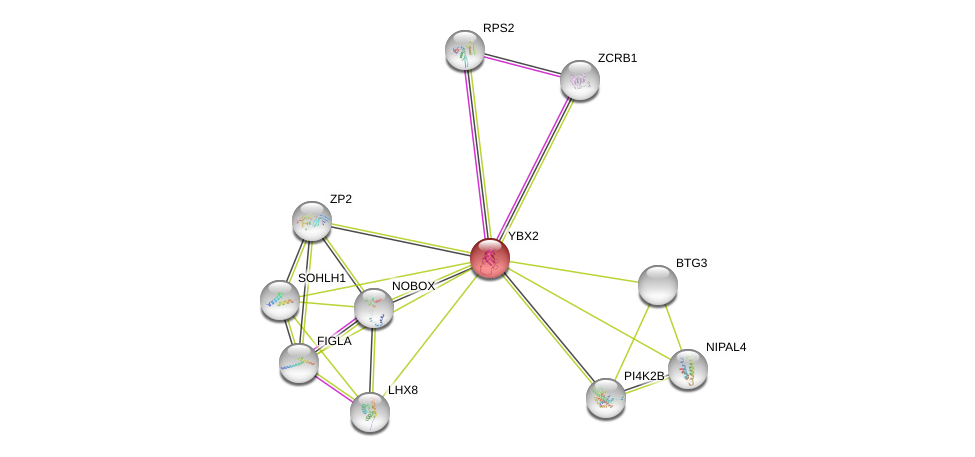
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000006047 | YBX2 | 54 | 85.204 | ENSAMEG00000016564 | YBX2 | 87 | 84.896 | Ailuropoda_melanoleuca |
| ENSG00000006047 | YBX2 | 99 | 87.730 | ENSANAG00000019540 | YBX2 | 86 | 88.926 | Aotus_nancymaae |
| ENSG00000006047 | YBX2 | 99 | 87.117 | ENSBTAG00000009126 | YBX2 | 84 | 89.935 | Bos_taurus |
| ENSG00000006047 | YBX2 | 99 | 84.049 | ENSCJAG00000042146 | - | 97 | 86.429 | Callithrix_jacchus |
| ENSG00000006047 | YBX2 | 99 | 83.436 | ENSCJAG00000016581 | - | 84 | 87.662 | Callithrix_jacchus |
| ENSG00000006047 | YBX2 | 95 | 63.462 | ENSCAFG00000005162 | - | 89 | 72.374 | Canis_familiaris |
| ENSG00000006047 | YBX2 | 99 | 84.756 | ENSCAFG00000016128 | - | 84 | 88.997 | Canis_familiaris |
| ENSG00000006047 | YBX2 | 99 | 87.117 | ENSCAFG00020008884 | YBX2 | 84 | 85.669 | Canis_lupus_dingo |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSCHIG00000016407 | YBX2 | 84 | 89.610 | Capra_hircus |
| ENSG00000006047 | YBX2 | 99 | 85.276 | ENSTSYG00000030765 | YBX2 | 92 | 83.571 | Carlito_syrichta |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSCAPG00000011432 | YBX2 | 100 | 89.011 | Cavia_aperea |
| ENSG00000006047 | YBX2 | 99 | 86.420 | ENSCPOG00000009543 | YBX2 | 96 | 88.768 | Cavia_porcellus |
| ENSG00000006047 | YBX2 | 99 | 88.957 | ENSCCAG00000025671 | YBX2 | 83 | 90.909 | Cebus_capucinus |
| ENSG00000006047 | YBX2 | 99 | 91.358 | ENSCATG00000036919 | YBX2 | 84 | 92.532 | Cercocebus_atys |
| ENSG00000006047 | YBX2 | 99 | 87.117 | ENSCLAG00000010306 | YBX2 | 96 | 89.493 | Chinchilla_lanigera |
| ENSG00000006047 | YBX2 | 99 | 91.358 | ENSCSAG00000011857 | YBX2 | 84 | 92.532 | Chlorocebus_sabaeus |
| ENSG00000006047 | YBX2 | 72 | 64.662 | ENSCPBG00000022288 | YBX2 | 83 | 67.279 | Chrysemys_picta_bellii |
| ENSG00000006047 | YBX2 | 99 | 89.571 | ENSCANG00000007129 | YBX2 | 97 | 91.241 | Colobus_angolensis_palliatus |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSCGRG00001000751 | Ybx2 | 96 | 88.768 | Cricetulus_griseus_chok1gshd |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSCGRG00000007683 | Ybx2 | 99 | 88.768 | Cricetulus_griseus_crigri |
| ENSG00000006047 | YBX2 | 99 | 84.049 | ENSDNOG00000013746 | YBX2 | 97 | 77.124 | Dasypus_novemcinctus |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSDORG00000010882 | Ybx2 | 95 | 89.051 | Dipodomys_ordii |
| ENSG00000006047 | YBX2 | 56 | 47.143 | ENSEBUG00000009872 | YBX2 | 60 | 46.766 | Eptatretus_burgeri |
| ENSG00000006047 | YBX2 | 95 | 85.806 | ENSEASG00005018928 | YBX2 | 83 | 85.000 | Equus_asinus_asinus |
| ENSG00000006047 | YBX2 | 99 | 82.317 | ENSECAG00000009996 | YBX2 | 79 | 83.172 | Equus_caballus |
| ENSG00000006047 | YBX2 | 99 | 87.730 | ENSFCAG00000018097 | YBX2 | 85 | 89.610 | Felis_catus |
| ENSG00000006047 | YBX2 | 68 | 64.542 | ENSGAGG00000016159 | YBX2 | 87 | 70.518 | Gopherus_agassizii |
| ENSG00000006047 | YBX2 | 100 | 96.986 | ENSGGOG00000007399 | YBX2 | 100 | 96.986 | Gorilla_gorilla |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSHGLG00000014186 | YBX2 | 84 | 89.286 | Heterocephalus_glaber_female |
| ENSG00000006047 | YBX2 | 99 | 87.730 | ENSSTOG00000008630 | YBX2 | 99 | 89.493 | Ictidomys_tridecemlineatus |
| ENSG00000006047 | YBX2 | 99 | 87.117 | ENSJJAG00000013072 | Ybx2 | 96 | 89.493 | Jaculus_jaculus |
| ENSG00000006047 | YBX2 | 99 | 87.730 | ENSLAFG00000014006 | YBX2 | 84 | 90.584 | Loxodonta_africana |
| ENSG00000006047 | YBX2 | 99 | 91.358 | ENSMFAG00000039921 | YBX2 | 84 | 92.532 | Macaca_fascicularis |
| ENSG00000006047 | YBX2 | 99 | 91.358 | ENSMNEG00000045221 | YBX2 | 84 | 92.532 | Macaca_nemestrina |
| ENSG00000006047 | YBX2 | 99 | 90.798 | ENSMLEG00000039452 | YBX2 | 97 | 92.701 | Mandrillus_leucophaeus |
| ENSG00000006047 | YBX2 | 99 | 65.432 | ENSMLEG00000029266 | - | 84 | 73.239 | Mandrillus_leucophaeus |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSMAUG00000022062 | Ybx2 | 84 | 89.610 | Mesocricetus_auratus |
| ENSG00000006047 | YBX2 | 99 | 85.276 | ENSMICG00000012453 | YBX2 | 97 | 88.686 | Microcebus_murinus |
| ENSG00000006047 | YBX2 | 99 | 87.117 | ENSMOCG00000013958 | Ybx2 | 96 | 89.493 | Microtus_ochrogaster |
| ENSG00000006047 | YBX2 | 83 | 86.029 | ENSMODG00000017596 | YBX2 | 78 | 83.860 | Monodelphis_domestica |
| ENSG00000006047 | YBX2 | 99 | 87.654 | MGP_CAROLIEiJ_G0016606 | Ybx2 | 96 | 89.493 | Mus_caroli |
| ENSG00000006047 | YBX2 | 99 | 87.654 | ENSMUSG00000018554 | Ybx2 | 96 | 89.493 | Mus_musculus |
| ENSG00000006047 | YBX2 | 99 | 87.654 | MGP_PahariEiJ_G0017737 | Ybx2 | 96 | 89.493 | Mus_pahari |
| ENSG00000006047 | YBX2 | 99 | 87.654 | MGP_SPRETEiJ_G0017442 | Ybx2 | 96 | 89.493 | Mus_spretus |
| ENSG00000006047 | YBX2 | 99 | 78.395 | ENSMPUG00000010386 | YBX2 | 93 | 83.333 | Mustela_putorius_furo |
| ENSG00000006047 | YBX2 | 99 | 85.185 | ENSMLUG00000004686 | YBX2 | 90 | 82.143 | Myotis_lucifugus |
| ENSG00000006047 | YBX2 | 99 | 84.049 | ENSNGAG00000011707 | Ybx2 | 85 | 87.987 | Nannospalax_galili |
| ENSG00000006047 | YBX2 | 99 | 91.358 | ENSNLEG00000008063 | - | 100 | 93.464 | Nomascus_leucogenys |
| ENSG00000006047 | YBX2 | 97 | 81.761 | ENSMEUG00000006947 | YBX2 | 90 | 79.221 | Notamacropus_eugenii |
| ENSG00000006047 | YBX2 | 99 | 82.209 | ENSOPRG00000013773 | YBX2 | 100 | 85.135 | Ochotona_princeps |
| ENSG00000006047 | YBX2 | 99 | 85.276 | ENSODEG00000017530 | YBX2 | 96 | 88.406 | Octodon_degus |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSOCUG00000015064 | YBX2 | 95 | 80.263 | Oryctolagus_cuniculus |
| ENSG00000006047 | YBX2 | 99 | 85.276 | ENSOGAG00000004392 | YBX2 | 100 | 88.686 | Otolemur_garnettii |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSOARG00000011562 | YBX2 | 78 | 87.986 | Ovis_aries |
| ENSG00000006047 | YBX2 | 99 | 100.000 | ENSPPAG00000041892 | YBX2 | 100 | 99.336 | Pan_paniscus |
| ENSG00000006047 | YBX2 | 99 | 87.037 | ENSPPRG00000000782 | YBX2 | 85 | 90.909 | Panthera_pardus |
| ENSG00000006047 | YBX2 | 99 | 100.000 | ENSPTRG00000008673 | YBX2 | 100 | 98.630 | Pan_troglodytes |
| ENSG00000006047 | YBX2 | 99 | 100.000 | ENSPTRG00000051288 | - | 85 | 99.265 | Pan_troglodytes |
| ENSG00000006047 | YBX2 | 99 | 89.571 | ENSPANG00000024250 | YBX2 | 84 | 92.131 | Papio_anubis |
| ENSG00000006047 | YBX2 | 99 | 62.346 | ENSPANG00000032747 | - | 74 | 67.208 | Papio_anubis |
| ENSG00000006047 | YBX2 | 75 | 66.312 | ENSPSIG00000012364 | YBX2 | 99 | 70.922 | Pelodiscus_sinensis |
| ENSG00000006047 | YBX2 | 97 | 83.019 | ENSPCIG00000028819 | YBX2 | 85 | 82.449 | Phascolarctos_cinereus |
| ENSG00000006047 | YBX2 | 99 | 85.185 | ENSPCAG00000008019 | YBX2 | 84 | 88.636 | Procavia_capensis |
| ENSG00000006047 | YBX2 | 99 | 84.663 | ENSPCOG00000006030 | YBX2 | 84 | 88.636 | Propithecus_coquereli |
| ENSG00000006047 | YBX2 | 99 | 83.333 | ENSPVAG00000005050 | YBX2 | 84 | 78.317 | Pteropus_vampyrus |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSRNOG00000017117 | Ybx2 | 84 | 89.610 | Rattus_norvegicus |
| ENSG00000006047 | YBX2 | 99 | 90.798 | ENSRBIG00000040919 | YBX2 | 85 | 92.208 | Rhinopithecus_bieti |
| ENSG00000006047 | YBX2 | 99 | 90.798 | ENSRROG00000041728 | YBX2 | 99 | 81.518 | Rhinopithecus_roxellana |
| ENSG00000006047 | YBX2 | 99 | 88.957 | ENSSBOG00000029566 | YBX2 | 97 | 90.511 | Saimiri_boliviensis_boliviensis |
| ENSG00000006047 | YBX2 | 97 | 82.390 | ENSSHAG00000007786 | YBX2 | 95 | 78.758 | Sarcophilus_harrisii |
| ENSG00000006047 | YBX2 | 99 | 85.890 | ENSSARG00000000488 | YBX2 | 84 | 89.286 | Sorex_araneus |
| ENSG00000006047 | YBX2 | 82 | 58.940 | ENSSPUG00000018250 | YBX2 | 99 | 64.026 | Sphenodon_punctatus |
| ENSG00000006047 | YBX2 | 99 | 87.654 | ENSSSCG00000017938 | YBX2 | 84 | 89.935 | Sus_scrofa |
| ENSG00000006047 | YBX2 | 99 | 82.099 | ENSTBEG00000001266 | YBX2 | 89 | 86.601 | Tupaia_belangeri |
| ENSG00000006047 | YBX2 | 57 | 97.938 | ENSTTRG00000009780 | YBX2 | 56 | 97.938 | Tursiops_truncatus |
| ENSG00000006047 | YBX2 | 99 | 86.503 | ENSUAMG00000014725 | YBX2 | 100 | 83.088 | Ursus_americanus |
| ENSG00000006047 | YBX2 | 85 | 87.857 | ENSUMAG00000015222 | YBX2 | 89 | 83.200 | Ursus_maritimus |
| ENSG00000006047 | YBX2 | 99 | 62.963 | ENSVPAG00000003104 | - | 100 | 71.825 | Vicugna_pacos |
| ENSG00000006047 | YBX2 | 95 | 80.128 | ENSVVUG00000012111 | - | 77 | 79.377 | Vulpes_vulpes |
| ENSG00000006047 | YBX2 | 99 | 85.276 | ENSVVUG00000027830 | - | 100 | 83.150 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0001650 | fibrillar center | - | IDA | Component |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003723 | RNA binding | - | IEA | Function |
| GO:0005634 | nucleus | 10100484. | TAS | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0006366 | transcription by RNA polymerase II | 10100484. | TAS | Process |
| GO:0007283 | spermatogenesis | 10100484. | TAS | Process |
| GO:0009386 | translational attenuation | 10100484. | TAS | Process |
| GO:0048599 | oocyte development | - | IEA | Process |
| GO:0120162 | positive regulation of cold-induced thermogenesis | 28970281. | ISS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| LAML | |||
| LIHC | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| MESO | ||
| ACC | ||
| BRCA | ||
| ESCA | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| GBM | ||
| LGG |