
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000394673 | ZNF207-204 | 2124 | - | ENSP00000378168 | 463 (aa) | - | O43670 |
| ENST00000582165 | ZNF207-214 | 592 | - | ENSP00000462193 | 95 (aa) | - | J3KRW6 |
| ENST00000579416 | ZNF207-209 | 396 | - | - | - (aa) | - | - |
| ENST00000582705 | ZNF207-215 | 333 | - | - | - (aa) | - | - |
| ENST00000321233 | ZNF207-201 | 2283 | - | ENSP00000322777 | 478 (aa) | - | O43670 |
| ENST00000580759 | ZNF207-212 | 572 | - | ENSP00000461914 | 93 (aa) | - | J3KRB6 |
| ENST00000584416 | ZNF207-216 | 1092 | - | - | - (aa) | - | - |
| ENST00000342555 | ZNF207-202 | 1905 | - | ENSP00000340029 | 497 (aa) | - | X6R4W8 |
| ENST00000394670 | ZNF207-203 | 13781 | XM_017025019 | ENSP00000378165 | 494 (aa) | XP_016880508 | O43670 |
| ENST00000579634 | ZNF207-210 | 580 | - | ENSP00000463040 | 74 (aa) | - | J3KTL1 |
| ENST00000577324 | ZNF207-206 | 1005 | - | - | - (aa) | - | - |
| ENST00000394679 | ZNF207-205 | 847 | - | ENSP00000378172 | 198 (aa) | - | H0Y3M2 |
| ENST00000581531 | ZNF207-213 | 3560 | - | ENSP00000462284 | 341 (aa) | - | J3KS31 |
| ENST00000584696 | ZNF207-217 | 743 | - | - | - (aa) | - | - |
| ENST00000579810 | ZNF207-211 | 979 | - | - | - (aa) | - | - |
| ENST00000578918 | ZNF207-208 | 580 | - | ENSP00000464495 | 46 (aa) | - | J3QS27 |
| ENST00000577908 | ZNF207-207 | 4672 | - | ENSP00000464366 | 493 (aa) | - | J3QRS9 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000010244 | rs9896320 | 17 | 32362795 | A | Smoking initiation (ever regular vs never regular) (MTAG) | 30643251 | [0.0049-0.0092] unit decrease | 0.00705081 | EFO_0006527 |
| ENSG00000010244 | rs2344976 | 17 | 32358916 | C | Smoking initiation (ever regular vs never regular) | 30643251 | [0.01-0.0202] unit decrease | 0.015087738 | EFO_0006527 |
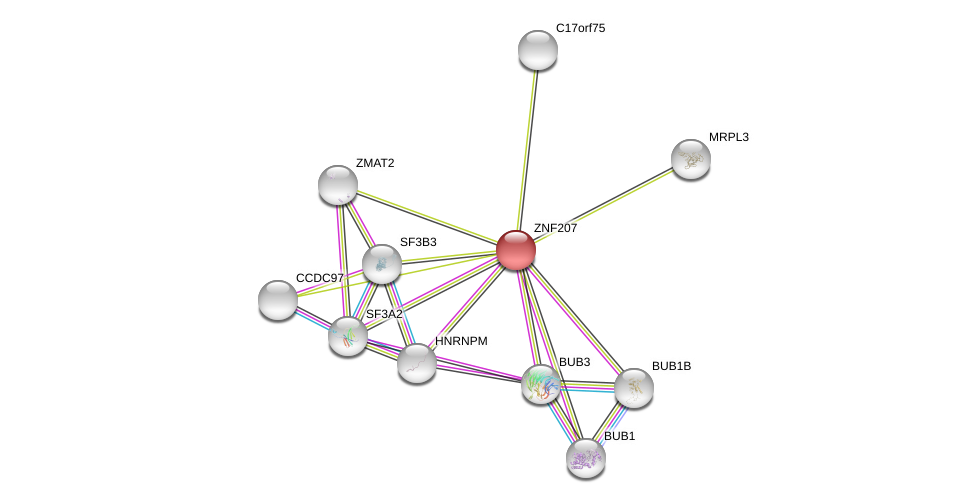
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000010244 | ZNF207 | 100 | 100.000 | ENSMUSG00000017421 | Zfp207 | 100 | 98.788 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000070 | mitotic sister chromatid segregation | 24462186.24462187. | IMP | Process |
| GO:0000776 | kinetochore | 21873635. | IBA | Component |
| GO:0000776 | kinetochore | 24462187. | IDA | Component |
| GO:0000776 | kinetochore | 24462186. | IMP | Component |
| GO:0000777 | condensed chromosome kinetochore | - | IEA | Component |
| GO:0001578 | microtubule bundle formation | - | ISS | Process |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003700 | DNA-binding transcription factor activity | 9799612. | NAS | Function |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005515 | protein binding | 24462186.24462187. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 24462186.24462187. | IDA | Component |
| GO:0005634 | nucleus | 9799612. | NAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0005819 | spindle | - | IEA | Component |
| GO:0005874 | microtubule | - | IEA | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 9799612. | NAS | Process |
| GO:0007094 | mitotic spindle assembly checkpoint | 21873635. | IBA | Process |
| GO:0007094 | mitotic spindle assembly checkpoint | 24462186.24462187. | IMP | Process |
| GO:0008017 | microtubule binding | 21873635. | IBA | Function |
| GO:0008017 | microtubule binding | 24462186.26388440. | IDA | Function |
| GO:0008201 | heparin binding | - | IEA | Function |
| GO:0008270 | zinc ion binding | 9799612. | NAS | Function |
| GO:0008608 | attachment of spindle microtubules to kinetochore | 21873635. | IBA | Process |
| GO:0008608 | attachment of spindle microtubules to kinetochore | 24462186.24462187. | IMP | Process |
| GO:0046785 | microtubule polymerization | - | ISS | Process |
| GO:0050821 | protein stabilization | 24462186.24462187. | IMP | Process |
| GO:0051301 | cell division | - | IEA | Process |
| GO:0051983 | regulation of chromosome segregation | 24462186.24462187. | IMP | Process |
| GO:0090307 | mitotic spindle assembly | 21873635. | IBA | Process |
| GO:0090307 | mitotic spindle assembly | 26388440. | IDA | Process |
| GO:1990047 | spindle matrix | 21873635. | IBA | Component |
| GO:1990047 | spindle matrix | 26388440. | IDA | Component |