
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000432309 | MDH1-205 | 844 | - | ENSP00000410073 | 222 (aa) | - | C9JF79 |
| ENST00000462944 | MDH1-209 | 634 | - | - | - (aa) | - | - |
| ENST00000436321 | MDH1-206 | 680 | - | ENSP00000394504 | 119 (aa) | - | C9JRL4 |
| ENST00000421012 | MDH1-204 | 573 | - | ENSP00000400937 | 87 (aa) | - | F8WFC2 |
| ENST00000484538 | MDH1-211 | 910 | - | - | - (aa) | - | - |
| ENST00000539945 | MDH1-215 | 1443 | - | ENSP00000438144 | 352 (aa) | - | P40925 |
| ENST00000233114 | MDH1-201 | 1650 | - | ENSP00000233114 | 334 (aa) | - | P40925 |
| ENST00000495083 | MDH1-214 | 552 | - | - | - (aa) | - | - |
| ENST00000485155 | MDH1-212 | 571 | - | - | - (aa) | - | - |
| ENST00000544381 | MDH1-216 | 1346 | - | ENSP00000446395 | 245 (aa) | - | P40925 |
| ENST00000442225 | MDH1-207 | 785 | - | ENSP00000399004 | 67 (aa) | - | C9IZI0 |
| ENST00000409908 | MDH1-203 | 700 | - | ENSP00000386743 | 169 (aa) | - | B8ZZ51 |
| ENST00000485781 | MDH1-213 | 893 | - | - | - (aa) | - | - |
| ENST00000409476 | MDH1-202 | 808 | - | ENSP00000386719 | 210 (aa) | - | B9A041 |
| ENST00000454035 | MDH1-208 | 575 | - | ENSP00000409027 | 114 (aa) | - | C9JLV6 |
| ENST00000472098 | MDH1-210 | 936 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00020 | Citrate cycle (TCA cycle) | KEGG |
| hsa00270 | Cysteine and methionine metabolism | KEGG |
| hsa00620 | Pyruvate metabolism | KEGG |
| hsa00630 | Glyoxylate and dicarboxylate metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
| hsa04964 | Proximal tubule bicarbonate reclamation | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000014641 | Blood Pressure | 3.4586055E-009 | - |
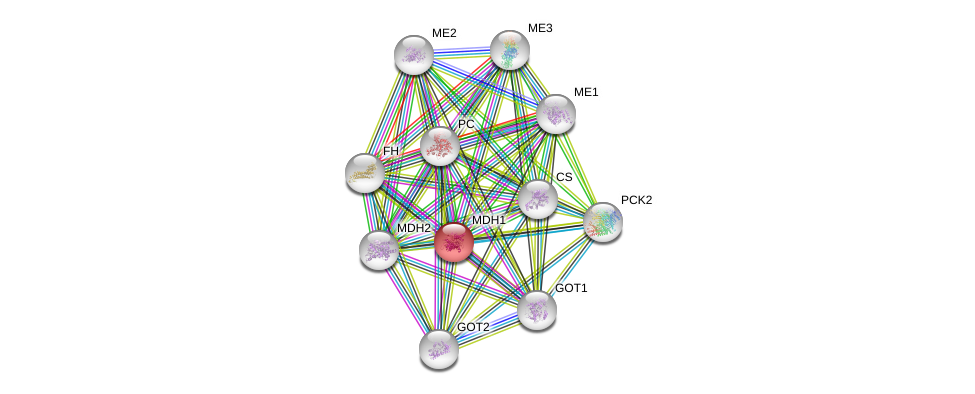
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004470 | malic enzyme activity | 8786100. | TAS | Function |
| GO:0005515 | protein binding | 21044950. | IPI | Function |
| GO:0005615 | extracellular space | 22664934. | HDA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006094 | gluconeogenesis | - | TAS | Process |
| GO:0006099 | tricarboxylic acid cycle | 21873635. | IBA | Process |
| GO:0006107 | oxaloacetate metabolic process | 21873635. | IBA | Process |
| GO:0006108 | malate metabolic process | 21873635. | IBA | Process |
| GO:0006734 | NADH metabolic process | 21873635. | IBA | Process |
| GO:0030060 | L-malate dehydrogenase activity | 21873635. | IBA | Function |
| GO:0047860 | diiodophenylpyruvate reductase activity | - | IEA | Function |
| GO:0051287 | NAD binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |