
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 1 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 2 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 3 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 4 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 5 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 6 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 7 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 8 | 9 |
| ENSP00000301310 | zf-C2H2 | PF00096.26 | 3.1e-54 | 9 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 1 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 2 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 3 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 4 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 5 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 6 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 7 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 8 | 9 |
| ENSP00000465619 | zf-C2H2 | PF00096.26 | 3.1e-54 | 9 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 1 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 2 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 3 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 4 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 5 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 6 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 7 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 8 | 9 |
| ENSP00000483414 | zf-C2H2 | PF00096.26 | 3.1e-54 | 9 | 9 |
| ENSP00000301310 | zf-met | PF12874.7 | 7.4e-08 | 1 | 1 |
| ENSP00000465619 | zf-met | PF12874.7 | 7.4e-08 | 1 | 1 |
| ENSP00000483414 | zf-met | PF12874.7 | 7.4e-08 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000301310 | ZNF582-201 | 2824 | - | ENSP00000301310 | 517 (aa) | - | Q96NG8 |
| ENST00000589895 | ZNF582-205 | 509 | - | ENSP00000465639 | 78 (aa) | - | K7EKI7 |
| ENST00000593145 | ZNF582-206 | 712 | - | ENSP00000466452 | 178 (aa) | - | A0A075B771 |
| ENST00000619584 | ZNF582-207 | 2297 | - | ENSP00000483414 | 517 (aa) | - | Q96NG8 |
| ENST00000589143 | ZNF582-204 | 625 | - | ENSP00000468679 | 103 (aa) | - | K7ESE7 |
| ENST00000586929 | ZNF582-202 | 2053 | - | ENSP00000465619 | 517 (aa) | - | Q96NG8 |
| ENST00000587778 | ZNF582-203 | 733 | - | ENSP00000466875 | 181 (aa) | - | A0A075B772 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05168 | Herpes simplex virus 1 infection | KEGG |
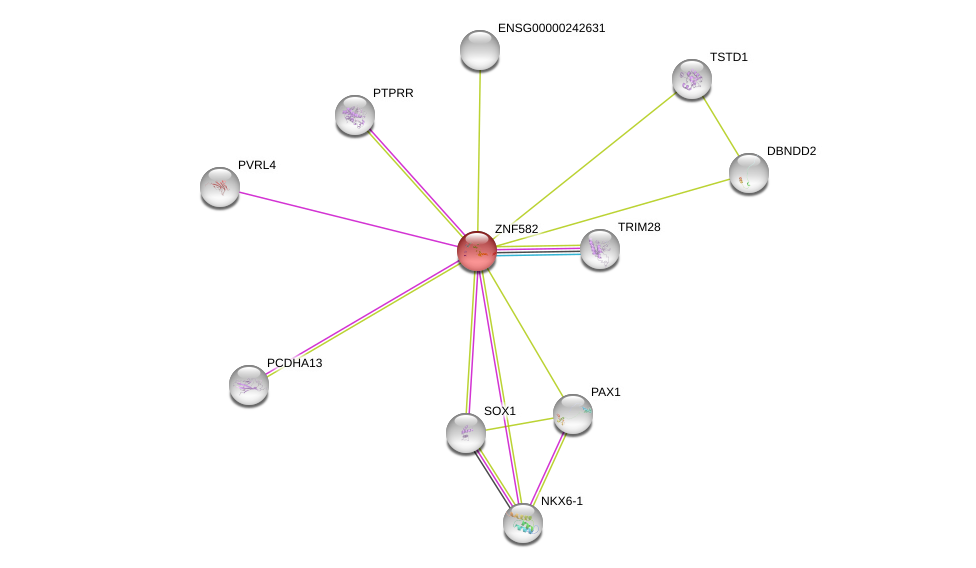
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000018869 | ZNF582 | 99 | 86.940 | ENSAMEG00000009560 | ZNF582 | 100 | 86.940 | Ailuropoda_melanoleuca |
| ENSG00000018869 | ZNF582 | 99 | 96.104 | ENSANAG00000026033 | ZNF582 | 94 | 94.971 | Aotus_nancymaae |
| ENSG00000018869 | ZNF582 | 100 | 87.621 | ENSBTAG00000030348 | ZNF582 | 100 | 87.621 | Bos_taurus |
| ENSG00000018869 | ZNF582 | 99 | 96.104 | ENSCJAG00000020283 | ZNF582 | 100 | 93.037 | Callithrix_jacchus |
| ENSG00000018869 | ZNF582 | 100 | 86.731 | ENSCHIG00000016463 | - | 100 | 86.796 | Capra_hircus |
| ENSG00000018869 | ZNF582 | 100 | 90.716 | ENSTSYG00000029281 | ZNF582 | 100 | 90.716 | Carlito_syrichta |
| ENSG00000018869 | ZNF582 | 99 | 84.416 | ENSCPOG00000032242 | - | 100 | 80.695 | Cavia_porcellus |
| ENSG00000018869 | ZNF582 | 99 | 96.104 | ENSCCAG00000025711 | ZNF582 | 100 | 94.971 | Cebus_capucinus |
| ENSG00000018869 | ZNF582 | 100 | 96.325 | ENSCATG00000036892 | ZNF582 | 94 | 96.325 | Cercocebus_atys |
| ENSG00000018869 | ZNF582 | 92 | 81.303 | ENSCLAG00000000758 | - | 88 | 81.303 | Chinchilla_lanigera |
| ENSG00000018869 | ZNF582 | 100 | 96.712 | ENSCSAG00000001670 | ZNF582 | 94 | 96.712 | Chlorocebus_sabaeus |
| ENSG00000018869 | ZNF582 | 92 | 85.645 | ENSCHOG00000012411 | ZNF582 | 89 | 85.645 | Choloepus_hoffmanni |
| ENSG00000018869 | ZNF582 | 99 | 97.403 | ENSCANG00000037320 | ZNF582 | 100 | 96.132 | Colobus_angolensis_palliatus |
| ENSG00000018869 | ZNF582 | 90 | 74.286 | ENSCGRG00001020646 | Gm3854 | 96 | 68.776 | Cricetulus_griseus_chok1gshd |
| ENSG00000018869 | ZNF582 | 90 | 74.286 | ENSCGRG00000008411 | Gm3854 | 96 | 68.776 | Cricetulus_griseus_crigri |
| ENSG00000018869 | ZNF582 | 100 | 89.942 | ENSEASG00005020473 | ZNF582 | 100 | 89.942 | Equus_asinus_asinus |
| ENSG00000018869 | ZNF582 | 91 | 89.407 | ENSECAG00000011892 | ZNF582 | 95 | 89.407 | Equus_caballus |
| ENSG00000018869 | ZNF582 | 100 | 100.000 | ENSGGOG00000003541 | ZNF582 | 100 | 99.226 | Gorilla_gorilla |
| ENSG00000018869 | ZNF582 | 98 | 85.743 | ENSHGLG00000009291 | - | 99 | 85.743 | Heterocephalus_glaber_female |
| ENSG00000018869 | ZNF582 | 98 | 85.743 | ENSHGLG00100007619 | ZNF582 | 99 | 85.743 | Heterocephalus_glaber_male |
| ENSG00000018869 | ZNF582 | 100 | 96.518 | ENSMFAG00000002728 | ZNF582 | 100 | 96.518 | Macaca_fascicularis |
| ENSG00000018869 | ZNF582 | 100 | 96.518 | ENSMMUG00000000362 | ZNF582 | 100 | 96.518 | Macaca_mulatta |
| ENSG00000018869 | ZNF582 | 100 | 96.132 | ENSMNEG00000028263 | ZNF582 | 99 | 96.195 | Macaca_nemestrina |
| ENSG00000018869 | ZNF582 | 100 | 96.132 | ENSMLEG00000030585 | ZNF582 | 100 | 96.132 | Mandrillus_leucophaeus |
| ENSG00000018869 | ZNF582 | 99 | 92.208 | ENSMICG00000012915 | ZNF582 | 100 | 91.103 | Microcebus_murinus |
| ENSG00000018869 | ZNF582 | 94 | 82.322 | ENSMPUG00000006660 | ZNF582 | 99 | 82.322 | Mustela_putorius_furo |
| ENSG00000018869 | ZNF582 | 94 | 79.630 | ENSNGAG00000000088 | Gm3854 | 99 | 79.630 | Nannospalax_galili |
| ENSG00000018869 | ZNF582 | 100 | 97.790 | ENSNLEG00000010794 | ZNF582 | 100 | 97.679 | Nomascus_leucogenys |
| ENSG00000018869 | ZNF582 | 94 | 75.103 | ENSODEG00000009980 | ZNF582 | 98 | 75.103 | Octodon_degus |
| ENSG00000018869 | ZNF582 | 99 | 85.380 | ENSOCUG00000029075 | ZNF582 | 100 | 85.380 | Oryctolagus_cuniculus |
| ENSG00000018869 | ZNF582 | 100 | 99.613 | ENSPPAG00000029277 | ZNF582 | 100 | 99.613 | Pan_paniscus |
| ENSG00000018869 | ZNF582 | 100 | 99.613 | ENSPTRG00000011539 | ZNF582 | 100 | 99.613 | Pan_troglodytes |
| ENSG00000018869 | ZNF582 | 100 | 96.518 | ENSPANG00000000398 | ZNF582 | 100 | 96.518 | Papio_anubis |
| ENSG00000018869 | ZNF582 | 99 | 100.000 | ENSPPYG00000010446 | ZNF582 | 91 | 97.679 | Pongo_abelii |
| ENSG00000018869 | ZNF582 | 92 | 78.947 | ENSPCAG00000000878 | ZNF582 | 90 | 78.947 | Procavia_capensis |
| ENSG00000018869 | ZNF582 | 99 | 94.805 | ENSPCOG00000014863 | ZNF582 | 100 | 93.230 | Propithecus_coquereli |
| ENSG00000018869 | ZNF582 | 100 | 96.325 | ENSRBIG00000042067 | ZNF582 | 94 | 96.325 | Rhinopithecus_bieti |
| ENSG00000018869 | ZNF582 | 100 | 96.132 | ENSRROG00000036529 | ZNF582 | 100 | 96.132 | Rhinopithecus_roxellana |
| ENSG00000018869 | ZNF582 | 99 | 96.104 | ENSSBOG00000024080 | ZNF582 | 94 | 94.391 | Saimiri_boliviensis_boliviensis |
| ENSG00000018869 | ZNF582 | 100 | 87.814 | ENSSSCG00000003322 | - | 100 | 87.814 | Sus_scrofa |
| ENSG00000018869 | ZNF582 | 100 | 89.362 | ENSTTRG00000011290 | ZNF582 | 91 | 89.362 | Tursiops_truncatus |
| ENSG00000018869 | ZNF582 | 88 | 86.374 | ENSUAMG00000010469 | ZNF582 | 99 | 86.374 | Ursus_americanus |
| ENSG00000018869 | ZNF582 | 88 | 86.593 | ENSUMAG00000024767 | - | 99 | 86.593 | Ursus_maritimus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| BRCA | |||
| CESC | |||
| DLBC | |||
| KIRP | |||
| LGG | |||
| PAAD | |||
| PRAD | |||
| READ | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| HNSC | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| LGG | ||
| LUAD |