
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 18279677 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000467772 | GCLM-203 | 625 | - | - | - (aa) | - | - |
| ENST00000615724 | GCLM-204 | 3008 | XM_011541261 | ENSP00000484507 | 252 (aa) | XP_011539563 | P48507 |
| ENST00000370238 | GCLM-201 | 4857 | XM_017001056 | ENSP00000359258 | 274 (aa) | XP_016856545 | P48507 |
| ENST00000462183 | GCLM-202 | 583 | - | - | - (aa) | - | - |
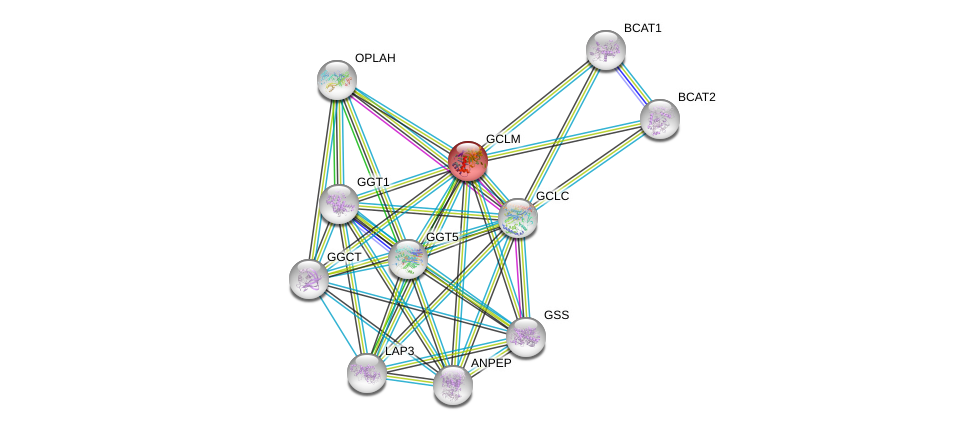
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000023909 | GCLM | 100 | 96.350 | ENSMUSG00000028124 | Gclm | 100 | 96.350 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004357 | glutamate-cysteine ligase activity | 9841880. | IDA | Function |
| GO:0004357 | glutamate-cysteine ligase activity | 16183645. | IMP | Function |
| GO:0005829 | cytosol | 9895302. | NAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006534 | cysteine metabolic process | - | IEA | Process |
| GO:0006536 | glutamate metabolic process | 9841880.9895302. | IDA | Process |
| GO:0006750 | glutathione biosynthetic process | 21873635. | IBA | Process |
| GO:0006750 | glutathione biosynthetic process | 10395918. | IDA | Process |
| GO:0006750 | glutathione biosynthetic process | - | IEA | Process |
| GO:0006750 | glutathione biosynthetic process | 12081989. | IMP | Process |
| GO:0006750 | glutathione biosynthetic process | - | TAS | Process |
| GO:0006979 | response to oxidative stress | 10395918. | IDA | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0007584 | response to nutrient | - | IEA | Process |
| GO:0008637 | apoptotic mitochondrial changes | - | IEA | Process |
| GO:0014823 | response to activity | - | IEA | Process |
| GO:0017109 | glutamate-cysteine ligase complex | 21873635. | IBA | Component |
| GO:0017109 | glutamate-cysteine ligase complex | 9841880. | IDA | Component |
| GO:0030234 | enzyme regulator activity | 21873635. | IBA | Function |
| GO:0035226 | glutamate-cysteine ligase catalytic subunit binding | 9675072.9841880. | IPI | Function |
| GO:0035229 | positive regulation of glutamate-cysteine ligase activity | 21873635. | IBA | Process |
| GO:0035729 | cellular response to hepatocyte growth factor stimulus | - | IEA | Process |
| GO:0035733 | hepatic stellate cell activation | - | IEA | Process |
| GO:0042493 | response to drug | 9895302. | IDA | Process |
| GO:0043524 | negative regulation of neuron apoptotic process | - | IEA | Process |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | - | IEA | Process |
| GO:0044752 | response to human chorionic gonadotropin | - | IEA | Process |
| GO:0046982 | protein heterodimerization activity | - | IEA | Function |
| GO:0050880 | regulation of blood vessel size | 12975258. | IMP | Process |
| GO:0051409 | response to nitrosative stress | - | IEA | Process |
| GO:0051900 | regulation of mitochondrial depolarization | - | IEA | Process |
| GO:0071333 | cellular response to glucose stimulus | - | IEA | Process |
| GO:0071372 | cellular response to follicle-stimulating hormone stimulus | - | IEA | Process |
| GO:0097069 | cellular response to thyroxine stimulus | - | IEA | Process |
| GO:1990830 | cellular response to leukemia inhibitory factor | - | IEA | Process |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BRCA | |||
| CESC | |||
| COAD | |||
| DLBC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| STAD | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| UCS | ||
| KIRP | ||
| COAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| GBM | ||
| LIHC | ||
| CHOL | ||
| LUAD |