
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000229446 | THRAP3_BCLAF1 | PF15440.6 | 4.3e-185 | 1 | 1 |
| ENSP00000376159 | THRAP3_BCLAF1 | PF15440.6 | 4.3e-185 | 1 | 1 |
| ENSP00000435441 | THRAP3_BCLAF1 | PF15440.6 | 4.5e-185 | 1 | 1 |
| ENSP00000434826 | THRAP3_BCLAF1 | PF15440.6 | 5.3e-185 | 1 | 1 |
| ENSP00000435210 | THRAP3_BCLAF1 | PF15440.6 | 5.5e-185 | 1 | 1 |
| ENSP00000487249 | THRAP3_BCLAF1 | PF15440.6 | 1.8e-184 | 1 | 1 |
| ENSP00000431734 | THRAP3_BCLAF1 | PF15440.6 | 1.3e-176 | 1 | 1 |
| ENSP00000436501 | THRAP3_BCLAF1 | PF15440.6 | 2.6e-135 | 1 | 2 |
| ENSP00000436501 | THRAP3_BCLAF1 | PF15440.6 | 2.6e-135 | 2 | 2 |
| ENSP00000437018 | Btz | PF09405.10 | 0.00081 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000530767 | BCLAF1-213 | 4186 | XM_017011529 | ENSP00000436501 | 747 (aa) | XP_016867018 | Q9NYF8 |
| ENST00000392348 | BCLAF1-202 | 2610 | - | ENSP00000376159 | 869 (aa) | - | Q9NYF8 |
| ENST00000640069 | BCLAF1-224 | 7719 | - | ENSP00000491530 | 703 (aa) | - | A0A1W2PQ43 |
| ENST00000534269 | BCLAF1-219 | 4239 | - | ENSP00000433822 | 467 (aa) | - | E9PJA7 |
| ENST00000534321 | BCLAF1-220 | 519 | - | - | - (aa) | - | - |
| ENST00000528229 | BCLAF1-208 | 573 | - | - | - (aa) | - | - |
| ENST00000531224 | BCLAF1-214 | 7263 | XM_017011532 | ENSP00000435210 | 920 (aa) | XP_016867021 | Q9NYF8 |
| ENST00000534762 | BCLAF1-221 | 744 | - | ENSP00000437018 | 187 (aa) | - | H0YF14 |
| ENST00000476194 | BCLAF1-203 | 630 | - | - | - (aa) | - | - |
| ENST00000529826 | BCLAF1-210 | 2423 | - | ENSP00000431734 | 725 (aa) | - | E9PK09 |
| ENST00000527759 | BCLAF1-207 | 4373 | - | ENSP00000434826 | 918 (aa) | - | Q9NYF8 |
| ENST00000530429 | BCLAF1-212 | 3167 | - | ENSP00000436142 | 750 (aa) | - | E9PQN2 |
| ENST00000532076 | BCLAF1-215 | 560 | - | - | - (aa) | - | - |
| ENST00000532384 | BCLAF1-216 | 3237 | - | ENSP00000433505 | 752 (aa) | - | E9PKI6 |
| ENST00000526228 | BCLAF1-204 | 572 | - | - | - (aa) | - | - |
| ENST00000533621 | BCLAF1-218 | 574 | - | ENSP00000436913 | 118 (aa) | - | H0YF00 |
| ENST00000353331 | BCLAF1-201 | 7110 | XM_017011531 | ENSP00000229446 | 869 (aa) | XP_016867020 | Q9NYF8 |
| ENST00000527613 | BCLAF1-206 | 5640 | - | ENSP00000436216 | 752 (aa) | - | E9PKI6 |
| ENST00000529522 | BCLAF1-209 | 1093 | - | - | - (aa) | - | - |
| ENST00000534792 | BCLAF1-222 | 552 | - | - | - (aa) | - | - |
| ENST00000527536 | BCLAF1-205 | 5371 | XM_005267237 | ENSP00000435441 | 871 (aa) | XP_005267294 | E9PK91 |
| ENST00000533422 | BCLAF1-217 | 692 | - | ENSP00000437333 | 126 (aa) | - | H0YF63 |
| ENST00000529917 | BCLAF1-211 | 1336 | - | - | - (aa) | - | - |
| ENST00000628517 | BCLAF1-223 | 2512 | - | ENSP00000487249 | 752 (aa) | - | E9PKI6 |
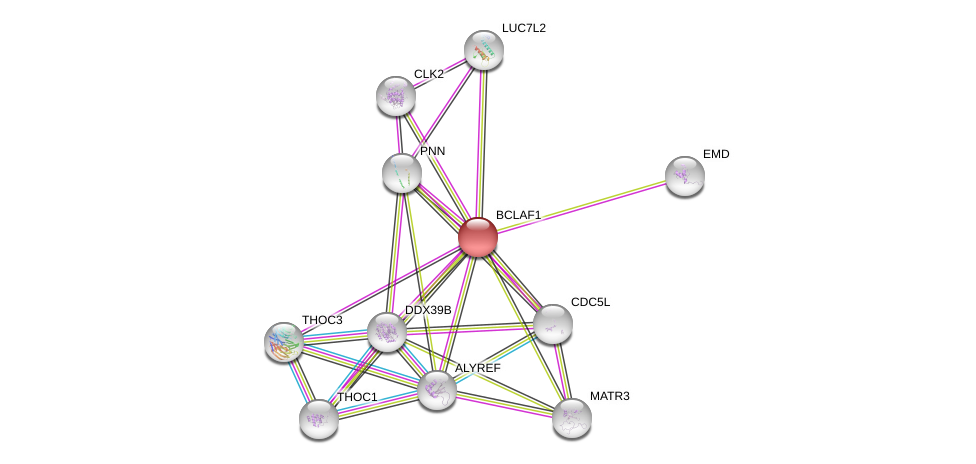
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000029363 | BCLAF1 | 100 | 97.802 | ENSAMEG00000009989 | BCLAF1 | 99 | 98.048 | Ailuropoda_melanoleuca |
| ENSG00000029363 | BCLAF1 | 62 | 54.237 | ENSACIG00000015331 | thrap3b | 50 | 41.741 | Amphilophus_citrinellus |
| ENSG00000029363 | BCLAF1 | 93 | 92.476 | ENSAPLG00000007570 | BCLAF1 | 94 | 89.516 | Anas_platyrhynchos |
| ENSG00000029363 | BCLAF1 | 93 | 88.862 | ENSACAG00000017011 | BCLAF1 | 94 | 84.943 | Anolis_carolinensis |
| ENSG00000029363 | BCLAF1 | 100 | 99.734 | ENSANAG00000035003 | BCLAF1 | 100 | 99.734 | Aotus_nancymaae |
| ENSG00000029363 | BCLAF1 | 100 | 98.817 | ENSBTAG00000007802 | BCLAF1 | 100 | 98.154 | Bos_taurus |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSCJAG00000008200 | BCLAF1 | 100 | 99.786 | Callithrix_jacchus |
| ENSG00000029363 | BCLAF1 | 100 | 98.371 | ENSCAFG00000000245 | BCLAF1 | 100 | 98.371 | Canis_familiaris |
| ENSG00000029363 | BCLAF1 | 93 | 98.361 | ENSCAFG00020022554 | BCLAF1 | 100 | 98.397 | Canis_lupus_dingo |
| ENSG00000029363 | BCLAF1 | 100 | 98.817 | ENSCHIG00000000525 | BCLAF1 | 100 | 98.046 | Capra_hircus |
| ENSG00000029363 | BCLAF1 | 65 | 49.194 | ENSTSYG00000011370 | - | 67 | 35.783 | Carlito_syrichta |
| ENSG00000029363 | BCLAF1 | 100 | 99.468 | ENSTSYG00000008161 | BCLAF1 | 100 | 99.468 | Carlito_syrichta |
| ENSG00000029363 | BCLAF1 | 98 | 98.475 | ENSCAPG00000009463 | - | 100 | 98.475 | Cavia_aperea |
| ENSG00000029363 | BCLAF1 | 100 | 98.043 | ENSCPOG00000008088 | BCLAF1 | 100 | 98.043 | Cavia_porcellus |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSCCAG00000032457 | BCLAF1 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000029363 | BCLAF1 | 100 | 99.867 | ENSCATG00000018862 | BCLAF1 | 100 | 99.867 | Cercocebus_atys |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSCATG00000038343 | - | 65 | 39.660 | Cercocebus_atys |
| ENSG00000029363 | BCLAF1 | 67 | 42.969 | ENSCATG00000035101 | - | 58 | 42.006 | Cercocebus_atys |
| ENSG00000029363 | BCLAF1 | 100 | 98.521 | ENSCLAG00000004401 | BCLAF1 | 100 | 98.046 | Chinchilla_lanigera |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSCSAG00000016867 | BCLAF1 | 100 | 99.657 | Chlorocebus_sabaeus |
| ENSG00000029363 | BCLAF1 | 60 | 55.224 | ENSCSAG00000001098 | THRAP3 | 65 | 39.876 | Chlorocebus_sabaeus |
| ENSG00000029363 | BCLAF1 | 93 | 91.990 | ENSCPBG00000017585 | BCLAF1 | 94 | 90.253 | Chrysemys_picta_bellii |
| ENSG00000029363 | BCLAF1 | 100 | 99.786 | ENSCANG00000043071 | BCLAF1 | 100 | 99.786 | Colobus_angolensis_palliatus |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSCANG00000043028 | THRAP3 | 65 | 39.660 | Colobus_angolensis_palliatus |
| ENSG00000029363 | BCLAF1 | 99 | 71.770 | ENSCANG00000003205 | - | 87 | 92.994 | Colobus_angolensis_palliatus |
| ENSG00000029363 | BCLAF1 | 99 | 96.800 | ENSCGRG00001024620 | - | 100 | 84.930 | Cricetulus_griseus_chok1gshd |
| ENSG00000029363 | BCLAF1 | 95 | 85.149 | ENSCGRG00001003851 | - | 93 | 75.143 | Cricetulus_griseus_chok1gshd |
| ENSG00000029363 | BCLAF1 | 100 | 87.167 | ENSCGRG00001015898 | - | 93 | 81.661 | Cricetulus_griseus_chok1gshd |
| ENSG00000029363 | BCLAF1 | 99 | 98.817 | ENSCGRG00000004873 | - | 93 | 87.500 | Cricetulus_griseus_crigri |
| ENSG00000029363 | BCLAF1 | 100 | 98.817 | ENSCGRG00000009622 | - | 100 | 97.503 | Cricetulus_griseus_crigri |
| ENSG00000029363 | BCLAF1 | 68 | 51.908 | ENSCVAG00000001547 | thrap3b | 64 | 35.726 | Cyprinodon_variegatus |
| ENSG00000029363 | BCLAF1 | 52 | 58.586 | ENSDARG00000098228 | thrap3b | 77 | 40.164 | Danio_rerio |
| ENSG00000029363 | BCLAF1 | 100 | 98.521 | ENSDNOG00000019473 | BCLAF1 | 100 | 97.477 | Dasypus_novemcinctus |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSDORG00000015144 | Bclaf1 | 100 | 97.937 | Dipodomys_ordii |
| ENSG00000029363 | BCLAF1 | 87 | 97.337 | ENSETEG00000015572 | BCLAF1 | 72 | 98.039 | Echinops_telfairi |
| ENSG00000029363 | BCLAF1 | 100 | 98.263 | ENSEASG00005011177 | BCLAF1 | 99 | 98.263 | Equus_asinus_asinus |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSECAG00000023692 | BCLAF1 | 100 | 98.371 | Equus_caballus |
| ENSG00000029363 | BCLAF1 | 52 | 53.731 | ENSEEUG00000004939 | THRAP3 | 50 | 47.870 | Erinaceus_europaeus |
| ENSG00000029363 | BCLAF1 | 93 | 98.587 | ENSEEUG00000008699 | BCLAF1 | 94 | 91.098 | Erinaceus_europaeus |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSFCAG00000027614 | BCLAF1 | 100 | 98.934 | Felis_catus |
| ENSG00000029363 | BCLAF1 | 93 | 92.494 | ENSFALG00000012928 | BCLAF1 | 94 | 88.953 | Ficedula_albicollis |
| ENSG00000029363 | BCLAF1 | 100 | 97.937 | ENSFDAG00000021270 | BCLAF1 | 100 | 97.937 | Fukomys_damarensis |
| ENSG00000029363 | BCLAF1 | 93 | 92.736 | ENSGALG00000013935 | BCLAF1 | 93 | 91.066 | Gallus_gallus |
| ENSG00000029363 | BCLAF1 | 93 | 91.748 | ENSGAGG00000023647 | BCLAF1 | 94 | 89.892 | Gopherus_agassizii |
| ENSG00000029363 | BCLAF1 | 94 | 88.316 | ENSGGOG00000042018 | - | 82 | 79.895 | Gorilla_gorilla |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSGGOG00000001824 | BCLAF1 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSHGLG00000001879 | BCLAF1 | 100 | 97.872 | Heterocephalus_glaber_female |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSHGLG00100008472 | BCLAF1 | 100 | 97.611 | Heterocephalus_glaber_male |
| ENSG00000029363 | BCLAF1 | 100 | 98.537 | ENSSTOG00000013977 | BCLAF1 | 100 | 98.537 | Ictidomys_tridecemlineatus |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSJJAG00000011803 | Bclaf1 | 100 | 97.477 | Jaculus_jaculus |
| ENSG00000029363 | BCLAF1 | 61 | 55.556 | ENSLBEG00000017803 | thrap3b | 50 | 41.871 | Labrus_bergylta |
| ENSG00000029363 | BCLAF1 | 100 | 74.822 | ENSLACG00000006728 | BCLAF1 | 83 | 62.883 | Latimeria_chalumnae |
| ENSG00000029363 | BCLAF1 | 61 | 59.130 | ENSLACG00000018229 | THRAP3 | 74 | 34.691 | Latimeria_chalumnae |
| ENSG00000029363 | BCLAF1 | 99 | 63.158 | ENSLOCG00000016208 | BCLAF1 | 74 | 65.625 | Lepisosteus_oculatus |
| ENSG00000029363 | BCLAF1 | 100 | 97.527 | ENSLAFG00000015982 | BCLAF1 | 99 | 97.939 | Loxodonta_africana |
| ENSG00000029363 | BCLAF1 | 58 | 60.870 | ENSLAFG00000009587 | THRAP3 | 65 | 39.692 | Loxodonta_africana |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSMFAG00000039754 | BCLAF1 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSMFAG00000040144 | - | 65 | 39.660 | Macaca_fascicularis |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSMMUG00000003710 | BCLAF1 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000029363 | BCLAF1 | 97 | 73.254 | ENSMMUG00000046285 | - | 78 | 91.463 | Macaca_mulatta |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSMMUG00000014622 | THRAP3 | 65 | 39.660 | Macaca_mulatta |
| ENSG00000029363 | BCLAF1 | 100 | 99.867 | ENSMNEG00000035970 | BCLAF1 | 100 | 99.867 | Macaca_nemestrina |
| ENSG00000029363 | BCLAF1 | 67 | 42.969 | ENSMNEG00000042630 | - | 71 | 38.992 | Macaca_nemestrina |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSMNEG00000018987 | - | 65 | 38.605 | Macaca_nemestrina |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSMLEG00000028924 | THRAP3 | 65 | 39.660 | Mandrillus_leucophaeus |
| ENSG00000029363 | BCLAF1 | 100 | 99.867 | ENSMLEG00000029438 | BCLAF1 | 100 | 99.867 | Mandrillus_leucophaeus |
| ENSG00000029363 | BCLAF1 | 93 | 92.476 | ENSMGAG00000013030 | BCLAF1 | 94 | 89.171 | Meleagris_gallopavo |
| ENSG00000029363 | BCLAF1 | 100 | 98.521 | ENSMAUG00000020380 | Bclaf1 | 100 | 96.851 | Mesocricetus_auratus |
| ENSG00000029363 | BCLAF1 | 100 | 99.358 | ENSMICG00000004946 | BCLAF1 | 100 | 99.358 | Microcebus_murinus |
| ENSG00000029363 | BCLAF1 | 63 | 57.724 | ENSMOCG00000004435 | - | 50 | 42.394 | Microtus_ochrogaster |
| ENSG00000029363 | BCLAF1 | 100 | 98.287 | ENSMOCG00000005885 | Bclaf1 | 100 | 98.287 | Microtus_ochrogaster |
| ENSG00000029363 | BCLAF1 | 65 | 48.062 | ENSMMOG00000018587 | thrap3a | 54 | 34.690 | Mola_mola |
| ENSG00000029363 | BCLAF1 | 99 | 87.081 | ENSMODG00000017447 | - | 92 | 93.595 | Monodelphis_domestica |
| ENSG00000029363 | BCLAF1 | 99 | 86.726 | ENSMODG00000005052 | - | 92 | 75.735 | Monodelphis_domestica |
| ENSG00000029363 | BCLAF1 | 99 | 99.200 | MGP_CAROLIEiJ_G0015157 | Bclaf1 | 100 | 94.007 | Mus_caroli |
| ENSG00000029363 | BCLAF1 | 52 | 53.731 | ENSMUSG00000043962 | Thrap3 | 97 | 45.522 | Mus_musculus |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSMUSG00000037608 | Bclaf1 | 100 | 94.178 | Mus_musculus |
| ENSG00000029363 | BCLAF1 | 52 | 53.731 | MGP_PahariEiJ_G0028802 | - | 97 | 44.776 | Mus_pahari |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | MGP_SPRETEiJ_G0015965 | Bclaf1 | 100 | 96.308 | Mus_spretus |
| ENSG00000029363 | BCLAF1 | 100 | 98.371 | ENSMPUG00000014831 | BCLAF1 | 100 | 98.371 | Mustela_putorius_furo |
| ENSG00000029363 | BCLAF1 | 100 | 98.521 | ENSMLUG00000002343 | BCLAF1 | 100 | 98.158 | Myotis_lucifugus |
| ENSG00000029363 | BCLAF1 | 99 | 99.200 | ENSNGAG00000016813 | Bclaf1 | 100 | 97.611 | Nannospalax_galili |
| ENSG00000029363 | BCLAF1 | 63 | 52.893 | ENSNBRG00000014237 | thrap3b | 50 | 41.425 | Neolamprologus_brichardi |
| ENSG00000029363 | BCLAF1 | 86 | 84.868 | ENSNLEG00000028319 | - | 78 | 83.906 | Nomascus_leucogenys |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSNLEG00000014796 | BCLAF1 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000029363 | BCLAF1 | 84 | 98.592 | ENSMEUG00000013217 | BCLAF1 | 85 | 98.611 | Notamacropus_eugenii |
| ENSG00000029363 | BCLAF1 | 77 | 97.222 | ENSOPRG00000012771 | BCLAF1 | 68 | 97.238 | Ochotona_princeps |
| ENSG00000029363 | BCLAF1 | 63 | 94.545 | ENSODEG00000000384 | - | 69 | 94.545 | Octodon_degus |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSODEG00000006806 | - | 100 | 97.394 | Octodon_degus |
| ENSG00000029363 | BCLAF1 | 99 | 86.538 | ENSOANG00000009444 | BCLAF1 | 93 | 94.431 | Ornithorhynchus_anatinus |
| ENSG00000029363 | BCLAF1 | 75 | 78.652 | ENSOCUG00000007115 | - | 86 | 73.684 | Oryctolagus_cuniculus |
| ENSG00000029363 | BCLAF1 | 100 | 97.937 | ENSOCUG00000016363 | - | 100 | 97.937 | Oryctolagus_cuniculus |
| ENSG00000029363 | BCLAF1 | 66 | 36.641 | ENSOMEG00000019164 | thrap3a | 50 | 35.202 | Oryzias_melastigma |
| ENSG00000029363 | BCLAF1 | 100 | 98.485 | ENSOGAG00000009724 | BCLAF1 | 99 | 98.478 | Otolemur_garnettii |
| ENSG00000029363 | BCLAF1 | 100 | 97.941 | ENSOARG00000000096 | BCLAF1 | 100 | 97.941 | Ovis_aries |
| ENSG00000029363 | BCLAF1 | 100 | 99.866 | ENSPPAG00000031645 | BCLAF1 | 100 | 99.866 | Pan_paniscus |
| ENSG00000029363 | BCLAF1 | 93 | 92.388 | ENSPPAG00000029536 | - | 93 | 91.468 | Pan_paniscus |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSPPRG00000023156 | - | 100 | 98.404 | Panthera_pardus |
| ENSG00000029363 | BCLAF1 | 84 | 82.828 | ENSPPRG00000003806 | - | 85 | 87.500 | Panthera_pardus |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSPTIG00000019385 | BCLAF1 | 100 | 98.271 | Panthera_tigris_altaica |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSPTRG00000018635 | BCLAF1 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000029363 | BCLAF1 | 96 | 92.459 | ENSPTRG00000049886 | - | 94 | 86.390 | Pan_troglodytes |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSPANG00000010532 | THRAP3 | 65 | 39.660 | Papio_anubis |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSPANG00000022182 | BCLAF1 | 100 | 100.000 | Papio_anubis |
| ENSG00000029363 | BCLAF1 | 78 | 87.931 | ENSPANG00000033308 | - | 84 | 87.709 | Papio_anubis |
| ENSG00000029363 | BCLAF1 | 93 | 91.525 | ENSPSIG00000007141 | BCLAF1 | 94 | 89.528 | Pelodiscus_sinensis |
| ENSG00000029363 | BCLAF1 | 92 | 93.881 | ENSPEMG00000012755 | - | 92 | 94.015 | Peromyscus_maniculatus_bairdii |
| ENSG00000029363 | BCLAF1 | 88 | 95.400 | ENSPCIG00000017609 | - | 87 | 94.915 | Phascolarctos_cinereus |
| ENSG00000029363 | BCLAF1 | 68 | 48.507 | ENSPFOG00000000525 | thrap3b | 50 | 39.140 | Poecilia_formosa |
| ENSG00000029363 | BCLAF1 | 68 | 48.507 | ENSPLAG00000016521 | thrap3b | 50 | 39.140 | Poecilia_latipinna |
| ENSG00000029363 | BCLAF1 | 68 | 48.872 | ENSPMEG00000009021 | thrap3b | 50 | 39.224 | Poecilia_mexicana |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSPPYG00000001544 | - | 71 | 46.535 | Pongo_abelii |
| ENSG00000029363 | BCLAF1 | 100 | 99.674 | ENSPPYG00000017032 | BCLAF1 | 100 | 99.674 | Pongo_abelii |
| ENSG00000029363 | BCLAF1 | 100 | 96.556 | ENSPCAG00000012422 | BCLAF1 | 100 | 96.938 | Procavia_capensis |
| ENSG00000029363 | BCLAF1 | 100 | 99.358 | ENSPCOG00000014683 | BCLAF1 | 100 | 99.358 | Propithecus_coquereli |
| ENSG00000029363 | BCLAF1 | 94 | 94.161 | ENSPVAG00000007233 | BCLAF1 | 100 | 96.000 | Pteropus_vampyrus |
| ENSG00000029363 | BCLAF1 | 63 | 53.333 | ENSPNYG00000003062 | thrap3b | 52 | 41.163 | Pundamilia_nyererei |
| ENSG00000029363 | BCLAF1 | 98 | 97.581 | ENSRNOG00000058050 | Mtfr2 | 94 | 95.617 | Rattus_norvegicus |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSRBIG00000029072 | BCLAF1 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSRBIG00000031284 | THRAP3 | 65 | 37.307 | Rhinopithecus_bieti |
| ENSG00000029363 | BCLAF1 | 78 | 83.333 | ENSRBIG00000044847 | - | 82 | 84.906 | Rhinopithecus_bieti |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSRROG00000040259 | BCLAF1 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000029363 | BCLAF1 | 59 | 54.839 | ENSRROG00000028592 | THRAP3 | 65 | 39.660 | Rhinopithecus_roxellana |
| ENSG00000029363 | BCLAF1 | 100 | 100.000 | ENSSBOG00000033973 | BCLAF1 | 100 | 99.867 | Saimiri_boliviensis_boliviensis |
| ENSG00000029363 | BCLAF1 | 93 | 95.415 | ENSSHAG00000010721 | BCLAF1 | 93 | 95.129 | Sarcophilus_harrisii |
| ENSG00000029363 | BCLAF1 | 98 | 51.659 | ENSSFOG00015017853 | bclaf1 | 74 | 40.061 | Scleropages_formosus |
| ENSG00000029363 | BCLAF1 | 100 | 98.521 | ENSSARG00000010591 | BCLAF1 | 85 | 98.521 | Sorex_araneus |
| ENSG00000029363 | BCLAF1 | 93 | 93.462 | ENSSPUG00000012092 | BCLAF1 | 94 | 90.219 | Sphenodon_punctatus |
| ENSG00000029363 | BCLAF1 | 65 | 51.908 | ENSSPAG00000009248 | thrap3b | 72 | 58.333 | Stegastes_partitus |
| ENSG00000029363 | BCLAF1 | 60 | 52.239 | ENSSSCG00000029954 | THRAP3 | 54 | 61.224 | Sus_scrofa |
| ENSG00000029363 | BCLAF1 | 100 | 98.789 | ENSSSCG00000004163 | BCLAF1 | 100 | 98.371 | Sus_scrofa |
| ENSG00000029363 | BCLAF1 | 93 | 92.476 | ENSTGUG00000010835 | BCLAF1 | 94 | 88.940 | Taeniopygia_guttata |
| ENSG00000029363 | BCLAF1 | 100 | 98.225 | ENSTBEG00000014886 | BCLAF1 | 94 | 91.098 | Tupaia_belangeri |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSTTRG00000002543 | BCLAF1 | 100 | 98.480 | Tursiops_truncatus |
| ENSG00000029363 | BCLAF1 | 99 | 99.112 | ENSUAMG00000006413 | BCLAF1 | 95 | 98.152 | Ursus_americanus |
| ENSG00000029363 | BCLAF1 | 100 | 99.112 | ENSUMAG00000024012 | BCLAF1 | 100 | 98.271 | Ursus_maritimus |
| ENSG00000029363 | BCLAF1 | 60 | 35.830 | ENSVPAG00000004656 | THRAP3 | 64 | 35.043 | Vicugna_pacos |
| ENSG00000029363 | BCLAF1 | 99 | 88.517 | ENSVPAG00000003924 | BCLAF1 | 84 | 94.952 | Vicugna_pacos |
| ENSG00000029363 | BCLAF1 | 99 | 98.291 | ENSVVUG00000000623 | BCLAF1 | 99 | 98.291 | Vulpes_vulpes |
| ENSG00000029363 | BCLAF1 | 91 | 68.085 | ENSXETG00000033794 | bclaf1 | 81 | 58.073 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003677 | DNA binding | 10330179. | IDA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 10837489.15009215.23541952. | IPI | Function |
| GO:0005634 | nucleus | 10330179. | IDA | Component |
| GO:0005654 | nucleoplasm | 24100041. | IDA | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0006915 | apoptotic process | 10330179. | TAS | Process |
| GO:0016607 | nuclear speck | 24100041. | IDA | Component |
| GO:0043065 | positive regulation of apoptotic process | 10330179. | IDA | Process |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 17938203. | IMP | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 10330179. | IDA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 10330179. | TAS | Process |
| GO:1990830 | cellular response to leukemia inhibitory factor | - | IEA | Process |
| GO:2000144 | positive regulation of DNA-templated transcription, initiation | 17938203. | IMP | Process |
| GO:2001022 | positive regulation of response to DNA damage stimulus | 17938203. | IMP | Process |
| GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 17938203. | IMP | Process |