
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000337926 | Surp | PF01805.20 | 2.6e-15 | 1 | 1 |
| ENSP00000389380 | Surp | PF01805.20 | 2.6e-15 | 1 | 1 |
| ENSP00000472286 | Surp | PF01805.20 | 2.6e-15 | 1 | 1 |
| ENSP00000472914 | Surp | PF01805.20 | 2.7e-15 | 1 | 1 |
| ENSP00000337926 | G-patch | PF01585.23 | 4.9e-14 | 1 | 1 |
| ENSP00000389380 | G-patch | PF01585.23 | 4.9e-14 | 1 | 1 |
| ENSP00000472286 | G-patch | PF01585.23 | 4.9e-14 | 1 | 1 |
| ENSP00000472914 | G-patch | PF01585.23 | 5e-14 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000597095 | SUGP2-207 | 536 | - | - | - (aa) | - | - |
| ENST00000452918 | SUGP2-203 | 6176 | - | ENSP00000389380 | 1082 (aa) | - | Q8IX01 |
| ENST00000597280 | SUGP2-209 | 814 | - | ENSP00000472878 | 65 (aa) | - | M0R2Y3 |
| ENST00000594773 | SUGP2-206 | 4169 | - | ENSP00000470915 | 1040 (aa) | - | Q8IX01 |
| ENST00000601879 | SUGP2-215 | 5546 | - | ENSP00000472286 | 1082 (aa) | - | Q8IX01 |
| ENST00000600239 | SUGP2-213 | 5565 | - | ENSP00000473187 | 841 (aa) | - | M0R3F6 |
| ENST00000594445 | SUGP2-205 | 732 | - | ENSP00000471042 | 111 (aa) | - | M0R065 |
| ENST00000600377 | SUGP2-214 | 4752 | XM_024451307 | ENSP00000472914 | 1096 (aa) | XP_024307075 | M0R2Z9 |
| ENST00000593795 | SUGP2-204 | 747 | - | ENSP00000472183 | 53 (aa) | - | M0R1Y3 |
| ENST00000337018 | SUGP2-202 | 3640 | - | ENSP00000337926 | 1082 (aa) | - | Q8IX01 |
| ENST00000597163 | SUGP2-208 | 423 | - | - | - (aa) | - | - |
| ENST00000330854 | SUGP2-201 | 3629 | - | ENSP00000332373 | 1030 (aa) | - | Q8IX01 |
| ENST00000598240 | SUGP2-211 | 3235 | - | ENSP00000472244 | 170 (aa) | - | M0R216 |
| ENST00000598863 | SUGP2-212 | 551 | - | - | - (aa) | - | - |
| ENST00000598202 | SUGP2-210 | 764 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000064607 | rs141292672 | 19 | 19020204 | ? | DNA methylation variation (age effect) | 30348214 | GO_0006306 |
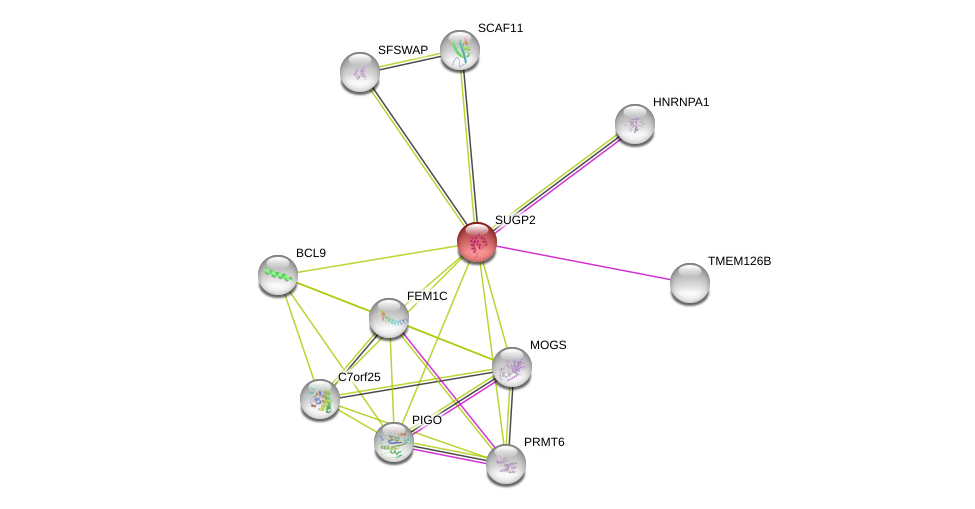
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000064607 | SUGP2 | 100 | 86.941 | ENSAMEG00000011613 | SUGP2 | 100 | 86.575 | Ailuropoda_melanoleuca |
| ENSG00000064607 | SUGP2 | 98 | 76.978 | ENSAPLG00000006469 | SUGP2 | 100 | 52.551 | Anas_platyrhynchos |
| ENSG00000064607 | SUGP2 | 100 | 96.396 | ENSANAG00000027159 | SUGP2 | 100 | 95.199 | Aotus_nancymaae |
| ENSG00000064607 | SUGP2 | 100 | 92.958 | ENSBTAG00000013288 | SUGP2 | 94 | 92.958 | Bos_taurus |
| ENSG00000064607 | SUGP2 | 100 | 97.297 | ENSCJAG00000019714 | SUGP2 | 100 | 95.568 | Callithrix_jacchus |
| ENSG00000064607 | SUGP2 | 100 | 87.281 | ENSCAFG00000014431 | SUGP2 | 100 | 87.281 | Canis_familiaris |
| ENSG00000064607 | SUGP2 | 100 | 87.061 | ENSCAFG00020012392 | SUGP2 | 100 | 87.056 | Canis_lupus_dingo |
| ENSG00000064607 | SUGP2 | 100 | 92.254 | ENSCHIG00000000571 | SUGP2 | 100 | 85.305 | Capra_hircus |
| ENSG00000064607 | SUGP2 | 100 | 89.246 | ENSTSYG00000027045 | SUGP2 | 100 | 89.020 | Carlito_syrichta |
| ENSG00000064607 | SUGP2 | 100 | 75.556 | ENSCAPG00000017450 | SUGP2 | 100 | 72.313 | Cavia_aperea |
| ENSG00000064607 | SUGP2 | 100 | 75.556 | ENSCPOG00000006372 | SUGP2 | 100 | 73.455 | Cavia_porcellus |
| ENSG00000064607 | SUGP2 | 100 | 96.396 | ENSCCAG00000023178 | SUGP2 | 100 | 96.214 | Cebus_capucinus |
| ENSG00000064607 | SUGP2 | 100 | 99.099 | ENSCATG00000029755 | SUGP2 | 100 | 97.786 | Cercocebus_atys |
| ENSG00000064607 | SUGP2 | 100 | 80.240 | ENSCLAG00000011375 | SUGP2 | 100 | 76.658 | Chinchilla_lanigera |
| ENSG00000064607 | SUGP2 | 100 | 99.099 | ENSCSAG00000005226 | SUGP2 | 100 | 97.970 | Chlorocebus_sabaeus |
| ENSG00000064607 | SUGP2 | 96 | 79.167 | ENSCPBG00000012487 | SUGP2 | 92 | 80.952 | Chrysemys_picta_bellii |
| ENSG00000064607 | SUGP2 | 100 | 97.297 | ENSCANG00000040758 | SUGP2 | 100 | 97.240 | Colobus_angolensis_palliatus |
| ENSG00000064607 | SUGP2 | 100 | 77.901 | ENSCGRG00001018264 | Sugp2 | 98 | 77.901 | Cricetulus_griseus_chok1gshd |
| ENSG00000064607 | SUGP2 | 100 | 77.901 | ENSCGRG00000009524 | Sugp2 | 98 | 77.901 | Cricetulus_griseus_crigri |
| ENSG00000064607 | SUGP2 | 75 | 77.165 | ENSDNOG00000037751 | - | 99 | 71.292 | Dasypus_novemcinctus |
| ENSG00000064607 | SUGP2 | 96 | 68.750 | ENSDORG00000011519 | Sugp2 | 100 | 69.164 | Dipodomys_ordii |
| ENSG00000064607 | SUGP2 | 99 | 70.065 | ENSETEG00000010805 | SUGP2 | 99 | 70.343 | Echinops_telfairi |
| ENSG00000064607 | SUGP2 | 100 | 86.517 | ENSEASG00005006565 | SUGP2 | 100 | 85.441 | Equus_asinus_asinus |
| ENSG00000064607 | SUGP2 | 100 | 86.839 | ENSECAG00000000129 | SUGP2 | 94 | 94.366 | Equus_caballus |
| ENSG00000064607 | SUGP2 | 100 | 87.158 | ENSFCAG00000009765 | SUGP2 | 100 | 87.432 | Felis_catus |
| ENSG00000064607 | SUGP2 | 99 | 77.273 | ENSFDAG00000021304 | SUGP2 | 99 | 75.386 | Fukomys_damarensis |
| ENSG00000064607 | SUGP2 | 92 | 81.000 | ENSGALG00000003047 | SUGP2 | 87 | 81.000 | Gallus_gallus |
| ENSG00000064607 | SUGP2 | 96 | 79.167 | ENSGAGG00000006784 | SUGP2 | 94 | 80.556 | Gopherus_agassizii |
| ENSG00000064607 | SUGP2 | 100 | 100.000 | ENSGGOG00000013540 | SUGP2 | 100 | 99.538 | Gorilla_gorilla |
| ENSG00000064607 | SUGP2 | 100 | 75.568 | ENSHGLG00000007497 | SUGP2 | 99 | 75.477 | Heterocephalus_glaber_female |
| ENSG00000064607 | SUGP2 | 97 | 75.908 | ENSHGLG00100006566 | SUGP2 | 99 | 74.672 | Heterocephalus_glaber_male |
| ENSG00000064607 | SUGP2 | 100 | 88.288 | ENSSTOG00000026887 | SUGP2 | 100 | 85.909 | Ictidomys_tridecemlineatus |
| ENSG00000064607 | SUGP2 | 100 | 75.676 | ENSJJAG00000005264 | Sugp2 | 100 | 74.497 | Jaculus_jaculus |
| ENSG00000064607 | SUGP2 | 85 | 63.235 | ENSLACG00000010085 | SUGP2 | 78 | 63.235 | Latimeria_chalumnae |
| ENSG00000064607 | SUGP2 | 100 | 78.105 | ENSLAFG00000018646 | SUGP2 | 100 | 78.473 | Loxodonta_africana |
| ENSG00000064607 | SUGP2 | 100 | 98.198 | ENSMFAG00000040352 | SUGP2 | 100 | 97.450 | Macaca_fascicularis |
| ENSG00000064607 | SUGP2 | 100 | 99.099 | ENSMMUG00000019665 | SUGP2 | 99 | 97.325 | Macaca_mulatta |
| ENSG00000064607 | SUGP2 | 100 | 99.099 | ENSMNEG00000043062 | SUGP2 | 100 | 97.632 | Macaca_nemestrina |
| ENSG00000064607 | SUGP2 | 96 | 97.603 | ENSMLEG00000036719 | SUGP2 | 100 | 97.603 | Mandrillus_leucophaeus |
| ENSG00000064607 | SUGP2 | 91 | 75.887 | ENSMGAG00000004496 | SUGP2 | 99 | 54.377 | Meleagris_gallopavo |
| ENSG00000064607 | SUGP2 | 100 | 77.348 | ENSMAUG00000011371 | Sugp2 | 100 | 73.035 | Mesocricetus_auratus |
| ENSG00000064607 | SUGP2 | 100 | 80.702 | ENSMOCG00000009704 | Sugp2 | 100 | 73.564 | Microtus_ochrogaster |
| ENSG00000064607 | SUGP2 | 100 | 60.374 | ENSMODG00000003992 | SUGP2 | 100 | 60.765 | Monodelphis_domestica |
| ENSG00000064607 | SUGP2 | 100 | 80.702 | MGP_CAROLIEiJ_G0031137 | Sugp2 | 100 | 72.678 | Mus_caroli |
| ENSG00000064607 | SUGP2 | 100 | 75.543 | ENSMUSG00000036054 | Sugp2 | 100 | 73.535 | Mus_musculus |
| ENSG00000064607 | SUGP2 | 100 | 79.532 | MGP_PahariEiJ_G0021602 | Sugp2 | 100 | 72.628 | Mus_pahari |
| ENSG00000064607 | SUGP2 | 100 | 76.087 | MGP_SPRETEiJ_G0032255 | Sugp2 | 100 | 73.718 | Mus_spretus |
| ENSG00000064607 | SUGP2 | 100 | 87.250 | ENSMPUG00000015478 | SUGP2 | 100 | 87.250 | Mustela_putorius_furo |
| ENSG00000064607 | SUGP2 | 100 | 84.147 | ENSMLUG00000012189 | SUGP2 | 100 | 84.147 | Myotis_lucifugus |
| ENSG00000064607 | SUGP2 | 100 | 79.444 | ENSNGAG00000005936 | Sugp2 | 100 | 74.702 | Nannospalax_galili |
| ENSG00000064607 | SUGP2 | 100 | 100.000 | ENSNLEG00000006162 | SUGP2 | 100 | 96.185 | Nomascus_leucogenys |
| ENSG00000064607 | SUGP2 | 86 | 83.688 | ENSMEUG00000011307 | SUGP2 | 96 | 63.774 | Notamacropus_eugenii |
| ENSG00000064607 | SUGP2 | 78 | 89.189 | ENSOPRG00000015993 | - | 84 | 69.156 | Ochotona_princeps |
| ENSG00000064607 | SUGP2 | 100 | 78.212 | ENSODEG00000012058 | SUGP2 | 100 | 75.708 | Octodon_degus |
| ENSG00000064607 | SUGP2 | 98 | 56.757 | ENSOANG00000004574 | SUGP2 | 99 | 56.645 | Ornithorhynchus_anatinus |
| ENSG00000064607 | SUGP2 | 100 | 90.991 | ENSOGAG00000024068 | SUGP2 | 100 | 89.770 | Otolemur_garnettii |
| ENSG00000064607 | SUGP2 | 100 | 92.958 | ENSOARG00000009811 | SUGP2 | 100 | 82.550 | Ovis_aries |
| ENSG00000064607 | SUGP2 | 96 | 99.424 | ENSPPAG00000043787 | SUGP2 | 98 | 99.424 | Pan_paniscus |
| ENSG00000064607 | SUGP2 | 100 | 87.511 | ENSPPRG00000010924 | SUGP2 | 100 | 87.603 | Panthera_pardus |
| ENSG00000064607 | SUGP2 | 100 | 86.888 | ENSPTIG00000012003 | SUGP2 | 100 | 86.981 | Panthera_tigris_altaica |
| ENSG00000064607 | SUGP2 | 100 | 100.000 | ENSPTRG00000010727 | SUGP2 | 100 | 99.361 | Pan_troglodytes |
| ENSG00000064607 | SUGP2 | 100 | 99.099 | ENSPANG00000004442 | SUGP2 | 100 | 97.632 | Papio_anubis |
| ENSG00000064607 | SUGP2 | 69 | 79.167 | ENSPSIG00000008256 | SUGP2 | 92 | 80.556 | Pelodiscus_sinensis |
| ENSG00000064607 | SUGP2 | 100 | 77.901 | ENSPEMG00000009700 | Sugp2 | 98 | 77.901 | Peromyscus_maniculatus_bairdii |
| ENSG00000064607 | SUGP2 | 100 | 59.946 | ENSPCIG00000000974 | SUGP2 | 99 | 60.978 | Phascolarctos_cinereus |
| ENSG00000064607 | SUGP2 | 100 | 100.000 | ENSPPYG00000009756 | SUGP2 | 100 | 98.244 | Pongo_abelii |
| ENSG00000064607 | SUGP2 | 100 | 89.935 | ENSPCOG00000016042 | SUGP2 | 100 | 89.718 | Propithecus_coquereli |
| ENSG00000064607 | SUGP2 | 74 | 84.921 | ENSPVAG00000005047 | SUGP2 | 74 | 87.739 | Pteropus_vampyrus |
| ENSG00000064607 | SUGP2 | 100 | 77.348 | ENSRNOG00000020269 | Sugp2 | 100 | 73.010 | Rattus_norvegicus |
| ENSG00000064607 | SUGP2 | 100 | 98.198 | ENSRBIG00000020354 | SUGP2 | 100 | 97.905 | Rhinopithecus_bieti |
| ENSG00000064607 | SUGP2 | 100 | 98.198 | ENSRROG00000029264 | SUGP2 | 100 | 97.970 | Rhinopithecus_roxellana |
| ENSG00000064607 | SUGP2 | 100 | 97.297 | ENSSBOG00000020629 | SUGP2 | 100 | 92.160 | Saimiri_boliviensis_boliviensis |
| ENSG00000064607 | SUGP2 | 100 | 60.536 | ENSSHAG00000011705 | SUGP2 | 100 | 61.552 | Sarcophilus_harrisii |
| ENSG00000064607 | SUGP2 | 100 | 74.354 | ENSSARG00000007461 | SUGP2 | 100 | 74.354 | Sorex_araneus |
| ENSG00000064607 | SUGP2 | 100 | 48.551 | ENSSPUG00000010736 | - | 100 | 49.139 | Sphenodon_punctatus |
| ENSG00000064607 | SUGP2 | 100 | 45.946 | ENSSPUG00000010778 | - | 100 | 46.335 | Sphenodon_punctatus |
| ENSG00000064607 | SUGP2 | 100 | 86.873 | ENSSSCG00000025839 | SUGP2 | 100 | 86.782 | Sus_scrofa |
| ENSG00000064607 | SUGP2 | 69 | 65.957 | ENSTGUG00000001088 | SUGP2 | 100 | 54.648 | Taeniopygia_guttata |
| ENSG00000064607 | SUGP2 | 100 | 92.254 | ENSTTRG00000006138 | SUGP2 | 100 | 86.606 | Tursiops_truncatus |
| ENSG00000064607 | SUGP2 | 100 | 87.158 | ENSUAMG00000007085 | SUGP2 | 100 | 86.794 | Ursus_americanus |
| ENSG00000064607 | SUGP2 | 100 | 83.698 | ENSUMAG00000006467 | SUGP2 | 100 | 83.424 | Ursus_maritimus |
| ENSG00000064607 | SUGP2 | 89 | 88.923 | ENSVPAG00000006943 | SUGP2 | 92 | 88.923 | Vicugna_pacos |
| ENSG00000064607 | SUGP2 | 100 | 87.079 | ENSVVUG00000007744 | SUGP2 | 100 | 85.755 | Vulpes_vulpes |
| ENSG00000064607 | SUGP2 | 65 | 37.573 | ENSXETG00000030450 | - | 98 | 38.305 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0006397 | mRNA processing | - | IEA | Process |
| GO:0008380 | RNA splicing | - | IEA | Process |
| GO:0016604 | nuclear body | - | IDA | Component |