
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000419860 | IBN_N | PF03810.19 | 1.5e-05 | 1 | 1 |
| ENSP00000419766 | IBN_N | PF03810.19 | 1.6e-05 | 1 | 1 |
| ENSP00000385938 | IBN_N | PF03810.19 | 2.2e-05 | 1 | 1 |
| ENSP00000420284 | IBN_N | PF03810.19 | 2.4e-05 | 1 | 1 |
| ENSP00000420079 | IBN_N | PF03810.19 | 2.8e-05 | 1 | 1 |
| ENSP00000420491 | IBN_N | PF03810.19 | 3.3e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000481455 | IPO5-216 | 555 | - | ENSP00000419766 | 119 (aa) | - | E7EX05 |
| ENST00000493122 | IPO5-224 | 651 | - | - | - (aa) | - | - |
| ENST00000631030 | IPO5-229 | 192 | - | ENSP00000487097 | 63 (aa) | - | F8WF30 |
| ENST00000473582 | IPO5-211 | 536 | - | ENSP00000420491 | 165 (aa) | - | C9JMV5 |
| ENST00000485433 | IPO5-219 | 524 | - | ENSP00000417721 | 75 (aa) | - | C9J875 |
| ENST00000491555 | IPO5-223 | 3688 | - | - | - (aa) | - | - |
| ENST00000479736 | IPO5-213 | 668 | - | - | - (aa) | - | - |
| ENST00000496368 | IPO5-227 | 542 | - | ENSP00000417162 | 98 (aa) | - | E7ESA1 |
| ENST00000493281 | IPO5-225 | 544 | - | ENSP00000419166 | 64 (aa) | - | C9JXE0 |
| ENST00000489058 | IPO5-220 | 522 | - | ENSP00000417993 | 50 (aa) | - | C9J583 |
| ENST00000490369 | IPO5-221 | 542 | - | ENSP00000419398 | 63 (aa) | - | F8WF30 |
| ENST00000421861 | IPO5-204 | 564 | - | ENSP00000408885 | 106 (aa) | - | E7ESZ1 |
| ENST00000460070 | IPO5-205 | 544 | - | ENSP00000420284 | 142 (aa) | - | E7EV12 |
| ENST00000490680 | IPO5-222 | 3419 | - | ENSP00000418393 | 1097 (aa) | - | O00410 |
| ENST00000481689 | IPO5-217 | 568 | - | ENSP00000420043 | 128 (aa) | - | E7ETV8 |
| ENST00000480611 | IPO5-214 | 591 | - | ENSP00000419860 | 117 (aa) | - | E7EWK4 |
| ENST00000463157 | IPO5-206 | 493 | - | ENSP00000420149 | 88 (aa) | - | C9JQT6 |
| ENST00000471898 | IPO5-210 | 556 | - | ENSP00000419797 | 74 (aa) | - | C9JZD8 |
| ENST00000482642 | IPO5-218 | 724 | - | - | - (aa) | - | - |
| ENST00000497270 | IPO5-228 | 536 | - | ENSP00000417647 | 126 (aa) | - | E7EQT5 |
| ENST00000468620 | IPO5-207 | 480 | - | - | - (aa) | - | - |
| ENST00000470493 | IPO5-209 | 548 | - | - | - (aa) | - | - |
| ENST00000403772 | IPO5-203 | 547 | - | ENSP00000385938 | 182 (aa) | - | H0Y3V4 |
| ENST00000475420 | IPO5-212 | 883 | - | ENSP00000420079 | 170 (aa) | - | E7ETV3 |
| ENST00000357602 | IPO5-202 | 4335 | - | ENSP00000350219 | 1097 (aa) | - | O00410 |
| ENST00000261574 | IPO5-201 | 6003 | XM_005254052 | ENSP00000261574 | 1115 (aa) | XP_005254109 | O00410 |
| ENST00000469360 | IPO5-208 | 3505 | - | ENSP00000419764 | 1099 (aa) | - | H0Y8C6 |
| ENST00000480641 | IPO5-215 | 543 | - | ENSP00000419003 | 77 (aa) | - | C9JZ53 |
| ENST00000493492 | IPO5-226 | 474 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000065150 | Bone Density | 5E-6 | 28181694 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000065150 | rs117023938 | 13 | 97989604 | T | Pediatric bone mineral density (hip) | 28181694 | [0.28-0.69] unit increase | 0.482788 | EFO_0007702 |
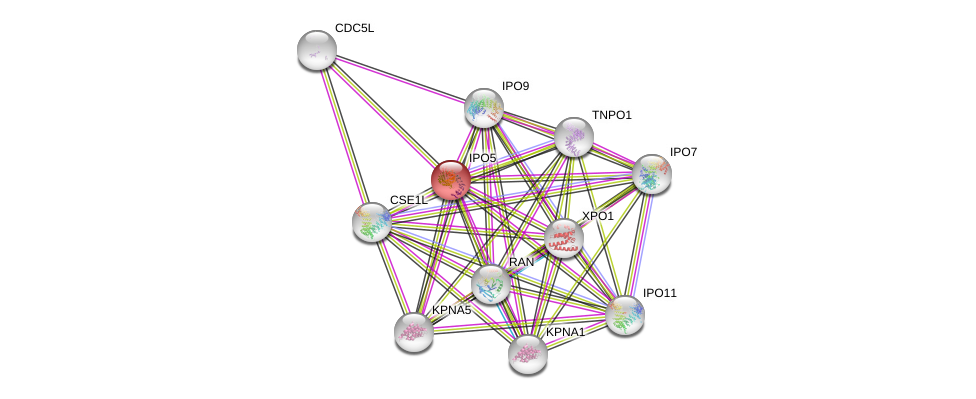
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000065150 | IPO5 | 99 | 93.613 | ENSACAG00000016245 | - | 99 | 93.613 | Anolis_carolinensis |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSANAG00000028954 | IPO5 | 97 | 99.458 | Aotus_nancymaae |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSCJAG00000004323 | IPO5 | 100 | 99.544 | Callithrix_jacchus |
| ENSG00000065150 | IPO5 | 99 | 96.007 | ENSTSYG00000005940 | IPO5 | 99 | 96.961 | Carlito_syrichta |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSCATG00000038175 | IPO5 | 100 | 98.394 | Cercocebus_atys |
| ENSG00000065150 | IPO5 | 87 | 88.073 | ENSCGRG00001014189 | - | 100 | 88.073 | Cricetulus_griseus_chok1gshd |
| ENSG00000065150 | IPO5 | 100 | 86.486 | ENSCGRG00001002524 | - | 89 | 87.037 | Cricetulus_griseus_chok1gshd |
| ENSG00000065150 | IPO5 | 100 | 93.506 | ENSETEG00000000152 | IPO5 | 93 | 92.715 | Echinops_telfairi |
| ENSG00000065150 | IPO5 | 99 | 89.570 | ENSLACG00000015338 | IPO5 | 99 | 89.551 | Latimeria_chalumnae |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSMFAG00000002597 | IPO5 | 100 | 99.821 | Macaca_fascicularis |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSMMUG00000013531 | IPO5 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSMNEG00000034763 | IPO5 | 100 | 98.394 | Macaca_nemestrina |
| ENSG00000065150 | IPO5 | 100 | 98.701 | ENSMLEG00000028889 | IPO5 | 99 | 98.389 | Mandrillus_leucophaeus |
| ENSG00000065150 | IPO5 | 100 | 98.701 | ENSMAUG00000016575 | - | 100 | 98.969 | Mesocricetus_auratus |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSMICG00000012711 | IPO5 | 100 | 96.825 | Microcebus_murinus |
| ENSG00000065150 | IPO5 | 98 | 96.465 | ENSMODG00000008520 | IPO5 | 97 | 94.555 | Monodelphis_domestica |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSNGAG00000014885 | Ipo5 | 100 | 96.727 | Nannospalax_galili |
| ENSG00000065150 | IPO5 | 99 | 100.000 | ENSMEUG00000002867 | IPO5 | 91 | 97.441 | Notamacropus_eugenii |
| ENSG00000065150 | IPO5 | 100 | 93.506 | ENSOPRG00000015494 | - | 100 | 87.295 | Ochotona_princeps |
| ENSG00000065150 | IPO5 | 100 | 96.354 | ENSODEG00000019128 | - | 100 | 96.354 | Octodon_degus |
| ENSG00000065150 | IPO5 | 100 | 94.805 | ENSODEG00000016371 | - | 100 | 92.069 | Octodon_degus |
| ENSG00000065150 | IPO5 | 100 | 97.403 | ENSODEG00000010941 | - | 100 | 91.682 | Octodon_degus |
| ENSG00000065150 | IPO5 | 91 | 80.000 | ENSOANG00000014256 | - | 83 | 75.610 | Ornithorhynchus_anatinus |
| ENSG00000065150 | IPO5 | 97 | 95.429 | ENSOANG00000005460 | - | 100 | 95.429 | Ornithorhynchus_anatinus |
| ENSG00000065150 | IPO5 | 100 | 97.403 | ENSOCUG00000003898 | IPO5 | 100 | 98.876 | Oryctolagus_cuniculus |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSPTRG00000005984 | IPO5 | 100 | 97.651 | Pan_troglodytes |
| ENSG00000065150 | IPO5 | 100 | 100.000 | ENSPANG00000009971 | IPO5 | 100 | 98.394 | Papio_anubis |
| ENSG00000065150 | IPO5 | 99 | 100.000 | ENSPCIG00000027635 | IPO5 | 100 | 97.091 | Phascolarctos_cinereus |
| ENSG00000065150 | IPO5 | 100 | 98.649 | ENSPCOG00000016473 | IPO5 | 100 | 96.997 | Propithecus_coquereli |
| ENSG00000065150 | IPO5 | 100 | 98.649 | ENSRNOG00000010989 | Ipo5 | 100 | 98.947 | Rattus_norvegicus |
| ENSG00000065150 | IPO5 | 100 | 98.701 | ENSRBIG00000036317 | IPO5 | 100 | 96.047 | Rhinopithecus_bieti |
| ENSG00000065150 | IPO5 | 100 | 97.403 | ENSSBOG00000028024 | IPO5 | 100 | 95.442 | Saimiri_boliviensis_boliviensis |
| ENSG00000065150 | IPO5 | 100 | 93.506 | ENSSHAG00000001752 | IPO5 | 99 | 93.601 | Sarcophilus_harrisii |
| ENSG00000065150 | IPO5 | 100 | 88.312 | ENSSPUG00000003786 | IPO5 | 100 | 93.413 | Sphenodon_punctatus |
| ENSG00000065150 | IPO5 | 99 | 90.876 | ENSTGUG00000011084 | IPO5 | 99 | 91.017 | Taeniopygia_guttata |
| ENSG00000065150 | IPO5 | 95 | 97.260 | ENSUAMG00000028286 | IPO5 | 99 | 98.171 | Ursus_americanus |
| ENSG00000065150 | IPO5 | 99 | 90.340 | ENSXETG00000023496 | ipo5 | 99 | 90.009 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005095 | GTPase inhibitor activity | 9271386. | TAS | Function |
| GO:0005515 | protein binding | 10799599.15282309.20562859.20828572.21044950.22730302. | IPI | Function |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005643 | nuclear pore | 9271386. | TAS | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0006607 | NLS-bearing protein import into nucleus | 21873635. | IBA | Process |
| GO:0006607 | NLS-bearing protein import into nucleus | - | ISS | Process |
| GO:0006610 | ribosomal protein import into nucleus | 21873635. | IBA | Process |
| GO:0006610 | ribosomal protein import into nucleus | 9687515. | IDA | Process |
| GO:0008139 | nuclear localization sequence binding | 21873635. | IBA | Function |
| GO:0008536 | Ran GTPase binding | - | IEA | Function |
| GO:0008565 | protein transporter activity | 21873635. | IBA | Function |
| GO:0008565 | protein transporter activity | - | ISS | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0031965 | nuclear membrane | 21873635. | IBA | Component |
| GO:0034260 | negative regulation of GTPase activity | - | IEA | Process |
| GO:0034399 | nuclear periphery | 21873635. | IBA | Component |
| GO:0042307 | positive regulation of protein import into nucleus | - | ISS | Process |
| GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 17491008. | TAS | Process |
| GO:0071230 | cellular response to amino acid stimulus | - | ISS | Process |