
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000484589 | CDK13-205 | 5166 | - | ENSP00000494206 | 456 (aa) | - | A0A2R8YD28 |
| ENST00000642592 | CDK13-209 | 1047 | - | ENSP00000494207 | 143 (aa) | - | A0A2R8YEB7 |
| ENST00000643868 | CDK13-213 | 1901 | - | ENSP00000495083 | 279 (aa) | - | A0A2R8YFE7 |
| ENST00000642213 | CDK13-208 | 1053 | - | - | - (aa) | - | - |
| ENST00000611390 | CDK13-206 | 2222 | - | ENSP00000484610 | 418 (aa) | - | A0A087X209 |
| ENST00000478563 | CDK13-204 | 904 | - | - | - (aa) | - | - |
| ENST00000642626 | CDK13-210 | 1150 | - | ENSP00000493853 | 288 (aa) | - | A0A2R8YCQ1 |
| ENST00000646437 | CDK13-220 | 772 | - | ENSP00000496618 | 257 (aa) | - | A0A2R8Y7Y0 |
| ENST00000642660 | CDK13-211 | 948 | - | - | - (aa) | - | - |
| ENST00000643915 | CDK13-214 | 3276 | - | ENSP00000496187 | 915 (aa) | - | A0A2R8YF61 |
| ENST00000340829 | CDK13-202 | 5455 | XM_017012751 | ENSP00000340557 | 1452 (aa) | XP_016868240 | Q14004 |
| ENST00000643859 | CDK13-212 | 3838 | - | ENSP00000496440 | 1099 (aa) | - | A0A2R8Y7W5 |
| ENST00000647453 | CDK13-221 | 2998 | - | - | - (aa) | - | - |
| ENST00000647518 | CDK13-222 | 4190 | - | - | - (aa) | - | - |
| ENST00000613626 | CDK13-207 | 3099 | - | ENSP00000480835 | 324 (aa) | - | Q9BVE2 |
| ENST00000645826 | CDK13-218 | 2250 | - | - | - (aa) | - | - |
| ENST00000465643 | CDK13-203 | 2394 | - | - | - (aa) | - | - |
| ENST00000646039 | CDK13-219 | 4466 | - | ENSP00000494168 | 1308 (aa) | - | A0A2R8Y4Z0 |
| ENST00000181839 | CDK13-201 | 10135 | XM_017012750 | ENSP00000181839 | 1512 (aa) | XP_016868239 | Q14004 |
| ENST00000644561 | CDK13-216 | 2576 | - | - | - (aa) | - | - |
| ENST00000644221 | CDK13-215 | 4511 | - | - | - (aa) | - | - |
| ENST00000645470 | CDK13-217 | 900 | - | ENSP00000495036 | 239 (aa) | - | A0A2R8Y644 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000065883 | Platelet Function Tests | 4.3500000E-006 | - |
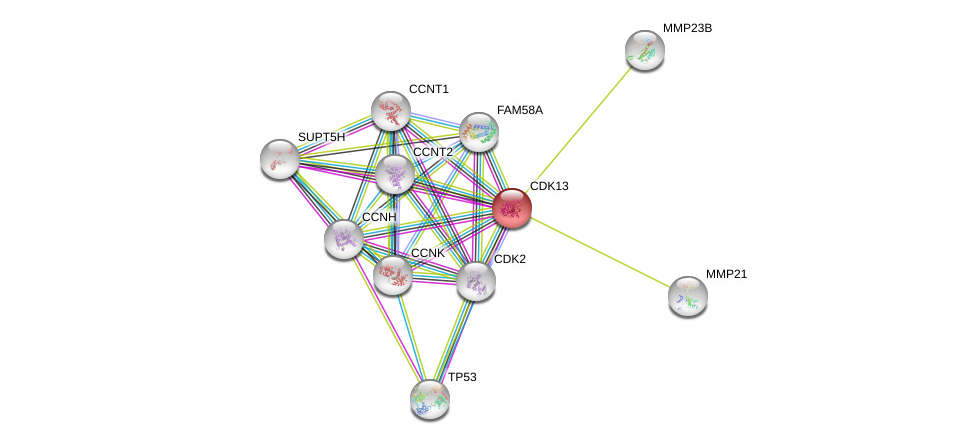
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000065883 | CDK13 | 100 | 99.163 | ENSMUSG00000041297 | Cdk13 | 100 | 98.276 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | 21873635. | IBA | Component |
| GO:0000380 | alternative mRNA splicing, via spliceosome | - | ISS | Process |
| GO:0000380 | alternative mRNA splicing, via spliceosome | 16721827. | TAS | Process |
| GO:0000790 | nuclear chromatin | 21873635. | IBA | Component |
| GO:0002945 | cyclin K-CDK13 complex | 22012619. | IPI | Component |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0004672 | protein kinase activity | 1731328. | TAS | Function |
| GO:0004693 | cyclin-dependent protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 16721827.26748711. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005615 | extracellular space | 22664934. | HDA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005694 | chromosome | 21873635. | IBA | Component |
| GO:0005794 | Golgi apparatus | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 21873635. | IBA | Process |
| GO:0006468 | protein phosphorylation | 21873635. | IBA | Process |
| GO:0007088 | regulation of mitotic nuclear division | 1731328. | TAS | Process |
| GO:0007275 | multicellular organism development | 1731328. | TAS | Process |
| GO:0008024 | cyclin/CDK positive transcription elongation factor complex | 21873635. | IBA | Component |
| GO:0008134 | transcription factor binding | 21873635. | IBA | Function |
| GO:0008284 | positive regulation of cell proliferation | 1731328. | TAS | Process |
| GO:0008353 | RNA polymerase II CTD heptapeptide repeat kinase activity | 21873635. | IBA | Function |
| GO:0008353 | RNA polymerase II CTD heptapeptide repeat kinase activity | 20952539. | IDA | Function |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016607 | nuclear speck | 16721827. | IDA | Component |
| GO:0019901 | protein kinase binding | - | IEA | Function |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | - | ISS | Component |
| GO:0030097 | hemopoiesis | 1731328. | IMP | Process |
| GO:0030332 | cyclin binding | 21873635. | IBA | Function |
| GO:0030332 | cyclin binding | 22012619. | IPI | Function |
| GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 21873635. | IBA | Process |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0044212 | transcription regulatory region DNA binding | 21873635. | IBA | Function |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain | 21873635. | IBA | Process |
| GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain | 20952539. | IDA | Process |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |
| GO:2000737 | negative regulation of stem cell differentiation | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| COAD | ||
| PAAD | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| UVM |