
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000495759 | ELP1-203 | 2840 | - | ENSP00000433514 | 346 (aa) | - | H0YDF3 |
| ENST00000467959 | ELP1-202 | 437 | - | - | - (aa) | - | - |
| ENST00000374647 | ELP1-201 | 5905 | XM_011519136 | ENSP00000363779 | 1332 (aa) | XP_011517438 | O95163 |
| ENST00000537196 | ELP1-204 | 3823 | - | ENSP00000439367 | 983 (aa) | - | F5H2T0 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000070061 | rs12353209 | 9 | 108890800 | ? | Myringotomy | 28928442 | [0.5-6.96] unit decrease | 3.7323 | EFO_0007904 |
| ENSG00000070061 | rs2275641 | 9 | 108934109 | C | Educational attainment (MTAG) | 30038396 | [0.0093-0.0147] unit increase | 0.012 | EFO_0004784 |
| ENSG00000070061 | rs2275641 | 9 | 108934109 | C | Highest math class taken (MTAG) | 30038396 | [0.0091-0.0157] unit increase | 0.0124 | EFO_0004875 |
| ENSG00000070061 | rs10979613 | 9 | 108925304 | T | Educational attainment (years of education) | 30038396 | [0.0088-0.0146] unit decrease | 0.0117 | EFO_0004784 |
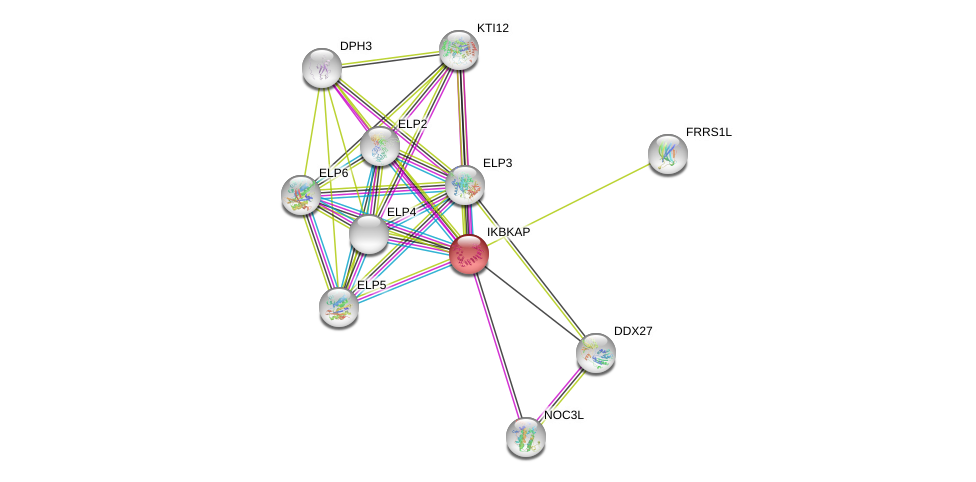
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000070061 | ELP1 | 100 | 80.899 | ENSMUSG00000028431 | Elp1 | 100 | 80.899 | Mus_musculus |
| ENSG00000070061 | ELP1 | 75 | 35.000 | YLR384C | IKI3 | 76 | 35.000 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000993 | RNA polymerase II complex binding | 11714725. | IDA | Function |
| GO:0002098 | tRNA wobble uridine modification | - | IEA | Process |
| GO:0005515 | protein binding | 11714725.15383276.19185337.21903422.22854966. | IPI | Function |
| GO:0005730 | nucleolus | 11714725. | IDA | Component |
| GO:0005737 | cytoplasm | 11714725.22854966. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | 11818576. | IDA | Process |
| GO:0006357 | regulation of transcription by RNA polymerase II | 11714725. | TAS | Process |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 11714725. | TAS | Process |
| GO:0006468 | protein phosphorylation | 9751059. | TAS | Process |
| GO:0006955 | immune response | 9751059. | TAS | Process |
| GO:0008023 | transcription elongation factor complex | 11714725. | IDA | Component |
| GO:0008607 | phosphorylase kinase regulator activity | 11818576. | IDA | Function |
| GO:0030335 | positive regulation of cell migration | - | ISS | Process |
| GO:0033588 | Elongator holoenzyme complex | 22854966. | IDA | Component |
| GO:0045859 | regulation of protein kinase activity | - | IEA | Process |
| GO:0065003 | protein-containing complex assembly | 9751059. | TAS | Process |