
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000453600 | ASNS-212 | 573 | - | ENSP00000408797 | 63 (aa) | - | C9JLN6 |
| ENST00000437657 | ASNS-207 | 745 | - | ENSP00000394242 | 162 (aa) | - | C9JT45 |
| ENST00000462436 | ASNS-215 | 698 | - | - | - (aa) | - | - |
| ENST00000437628 | ASNS-206 | 1638 | - | ENSP00000414379 | 478 (aa) | - | P08243 |
| ENST00000414884 | ASNS-204 | 584 | - | ENSP00000413797 | 28 (aa) | - | C9JLA3 |
| ENST00000448127 | ASNS-210 | 579 | - | ENSP00000402350 | 118 (aa) | - | C9JM09 |
| ENST00000454046 | ASNS-213 | 1947 | - | ENSP00000401651 | 401 (aa) | - | F8WEJ5 |
| ENST00000444334 | ASNS-209 | 1812 | - | ENSP00000406994 | 540 (aa) | - | P08243 |
| ENST00000455086 | ASNS-214 | 1588 | - | ENSP00000408472 | 478 (aa) | - | P08243 |
| ENST00000495255 | ASNS-217 | 1678 | - | - | - (aa) | - | - |
| ENST00000422745 | ASNS-205 | 1865 | - | ENSP00000414901 | 540 (aa) | - | P08243 |
| ENST00000487714 | ASNS-216 | 689 | - | - | - (aa) | - | - |
| ENST00000442734 | ASNS-208 | 828 | - | ENSP00000400422 | 214 (aa) | - | C9J057 |
| ENST00000175506 | ASNS-201 | 2356 | - | ENSP00000175506 | 561 (aa) | - | P08243 |
| ENST00000394308 | ASNS-202 | 1994 | - | ENSP00000377845 | 561 (aa) | - | P08243 |
| ENST00000394309 | ASNS-203 | 2289 | - | ENSP00000377846 | 561 (aa) | - | P08243 |
| ENST00000451771 | ASNS-211 | 562 | - | ENSP00000397802 | 32 (aa) | - | C9J605 |
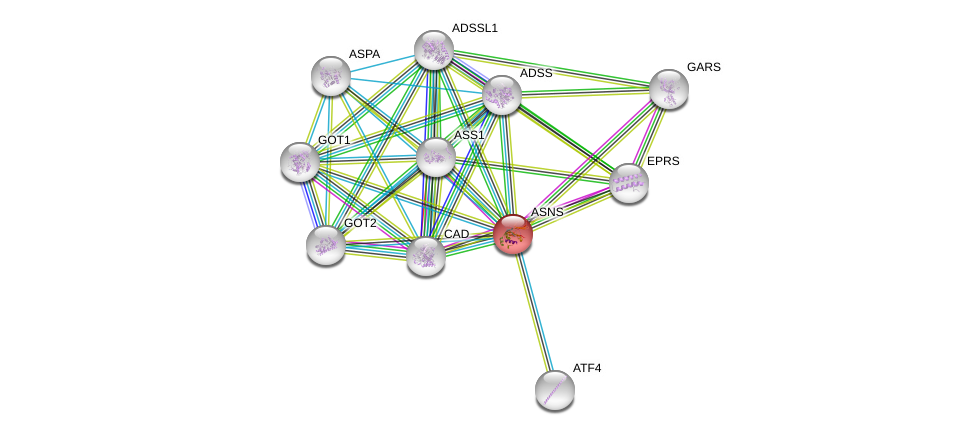
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001889 | liver development | - | IEA | Process |
| GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity | 21873635. | IBA | Function |
| GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity | 2564390.2569668.2573597.2886907.16023613. | IDA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006529 | asparagine biosynthetic process | 21873635. | IBA | Process |
| GO:0006529 | asparagine biosynthetic process | 2564390. | IC | Process |
| GO:0006529 | asparagine biosynthetic process | 2573597. | IDA | Process |
| GO:0006541 | glutamine metabolic process | - | IEA | Process |
| GO:0008652 | cellular amino acid biosynthetic process | - | TAS | Process |
| GO:0009416 | response to light stimulus | - | IEA | Process |
| GO:0009612 | response to mechanical stimulus | - | IEA | Process |
| GO:0009636 | response to toxic substance | - | IEA | Process |
| GO:0031427 | response to methotrexate | - | IEA | Process |
| GO:0032354 | response to follicle-stimulating hormone | - | IEA | Process |
| GO:0032870 | cellular response to hormone stimulus | - | IEA | Process |
| GO:0036499 | PERK-mediated unfolded protein response | - | TAS | Process |
| GO:0042149 | cellular response to glucose starvation | 10085239. | IDA | Process |
| GO:0042803 | protein homodimerization activity | 21873635. | IBA | Function |
| GO:0043066 | negative regulation of apoptotic process | 17409444. | IMP | Process |
| GO:0043200 | response to amino acid | - | IEA | Process |
| GO:0045931 | positive regulation of mitotic cell cycle | 2569668. | IDA | Process |
| GO:0048037 | cofactor binding | - | IEA | Function |
| GO:0070981 | L-asparagine biosynthetic process | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| DLBC | |||
| PAAD | |||
| PRAD | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| SARC | ||
| MESO | ||
| HNSC | ||
| PRAD | ||
| LUSC | ||
| KIRP | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG |