
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 26632259 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000477706 | HSD17B10-204 | 556 | - | - | - (aa) | - | - |
| ENST00000375304 | HSD17B10-203 | 916 | - | ENSP00000364453 | 252 (aa) | - | Q99714 |
| ENST00000495986 | HSD17B10-205 | 666 | - | - | - (aa) | - | - |
| ENST00000375298 | HSD17B10-202 | 823 | - | ENSP00000364447 | 169 (aa) | - | Q5H928 |
| ENST00000168216 | HSD17B10-201 | 960 | - | ENSP00000168216 | 261 (aa) | - | Q99714 |
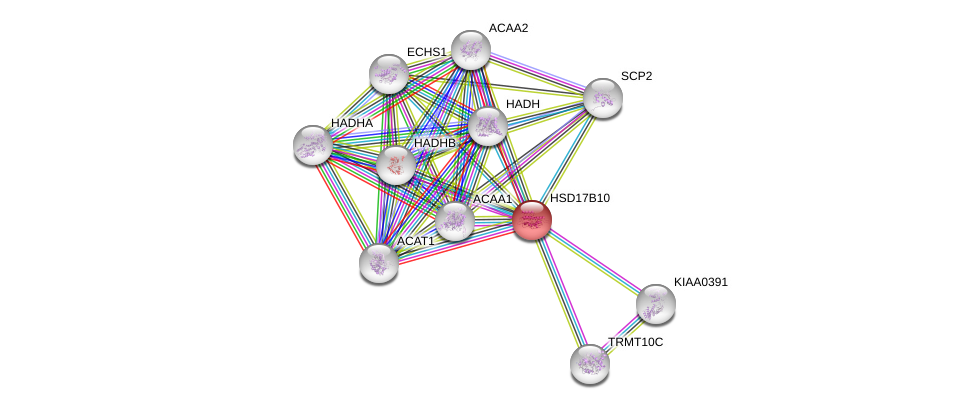
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000072506 | HSD17B10 | 97 | 68.898 | FBgn0021765 | scu | 100 | 75.397 | Drosophila_melanogaster |
| ENSG00000072506 | HSD17B10 | 100 | 87.739 | ENSMUSG00000025260 | Hsd17b10 | 100 | 87.739 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000049 | tRNA binding | 29040705. | IDA | Function |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity | 23042678.25925575.28888424. | IDA | Function |
| GO:0005515 | protein binding | 9338779.18984158.23042678.29040705.29128334. | IPI | Function |
| GO:0005737 | cytoplasm | 9338779. | TAS | Component |
| GO:0005739 | mitochondrion | 23042678. | IDA | Component |
| GO:0005739 | mitochondrion | - | ISS | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0005886 | plasma membrane | 9338779. | TAS | Component |
| GO:0006629 | lipid metabolic process | 9338779. | TAS | Process |
| GO:0007005 | mitochondrion organization | 20077426. | IMP | Process |
| GO:0008709 | cholate 7-alpha-dehydrogenase activity | 9338779. | TAS | Function |
| GO:0009083 | branched-chain amino acid catabolic process | - | TAS | Process |
| GO:0030283 | testosterone dehydrogenase [NAD(P)] activity | 28888424. | IDA | Function |
| GO:0030678 | mitochondrial ribonuclease P complex | 25925575.28888424. | IDA | Component |
| GO:0030678 | mitochondrial ribonuclease P complex | 23042678. | TAS | Component |
| GO:0035410 | dihydrotestosterone 17-beta-dehydrogenase activity | - | IEA | Function |
| GO:0047015 | 3-hydroxy-2-methylbutyryl-CoA dehydrogenase activity | - | IEA | Function |
| GO:0051289 | protein homotetramerization | 25925575. | IDA | Process |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |
| GO:0070901 | mitochondrial tRNA methylation | 25925575.28888424. | IDA | Process |
| GO:0070901 | mitochondrial tRNA methylation | - | TAS | Process |
| GO:0090646 | mitochondrial tRNA processing | - | TAS | Process |
| GO:0097745 | mitochondrial tRNA 5'-end processing | 24549042.25925575.28888424.29040705. | IDA | Process |
| GO:1990180 | mitochondrial tRNA 3'-end processing | 29040705. | IDA | Process |