
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000509564 | SDHA-211 | 630 | - | ENSP00000421911 | 101 (aa) | - | H0Y8S2 |
| ENST00000510361 | SDHA-213 | 2148 | XM_024446143 | ENSP00000427703 | 616 (aa) | XP_024301911 | P31040 |
| ENST00000507266 | SDHA-207 | 432 | - | - | - (aa) | - | - |
| ENST00000509082 | SDHA-209 | 434 | - | - | - (aa) | - | - |
| ENST00000512962 | SDHA-215 | 935 | - | - | - (aa) | - | - |
| ENST00000514233 | SDHA-217 | 558 | - | - | - (aa) | - | - |
| ENST00000505555 | SDHA-206 | 2230 | - | - | - (aa) | - | - |
| ENST00000617470 | SDHA-220 | 1952 | - | ENSP00000484230 | 519 (aa) | - | A0A087X1I3 |
| ENST00000509632 | SDHA-212 | 583 | - | ENSP00000425077 | 89 (aa) | - | D6REB7 |
| ENST00000511810 | SDHA-214 | 3001 | - | - | - (aa) | - | - |
| ENST00000515752 | SDHA-218 | 1809 | - | - | - (aa) | - | - |
| ENST00000514027 | SDHA-216 | 2209 | - | - | - (aa) | - | - |
| ENST00000504309 | SDHA-204 | 2067 | XM_011514073 | ENSP00000426514 | 583 (aa) | XP_011512375 | D6RFM5 |
| ENST00000503674 | SDHA-203 | 2426 | - | - | - (aa) | - | - |
| ENST00000502379 | SDHA-202 | 679 | - | - | - (aa) | - | - |
| ENST00000504824 | SDHA-205 | 1430 | - | - | - (aa) | - | - |
| ENST00000515815 | SDHA-219 | 704 | - | ENSP00000422404 | 126 (aa) | - | H0Y8X1 |
| ENST00000264932 | SDHA-201 | 2390 | XM_011514072 | ENSP00000264932 | 664 (aa) | XP_011512374 | P31040 |
| ENST00000507522 | SDHA-208 | 644 | - | - | - (aa) | - | - |
| ENST00000509420 | SDHA-210 | 580 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00020 | Citrate cycle (TCA cycle) | KEGG |
| hsa00190 | Oxidative phosphorylation | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
| hsa04714 | Thermogenesis | KEGG |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | KEGG |
| hsa05010 | Alzheimer disease | KEGG |
| hsa05012 | Parkinson disease | KEGG |
| hsa05016 | Huntington disease | KEGG |
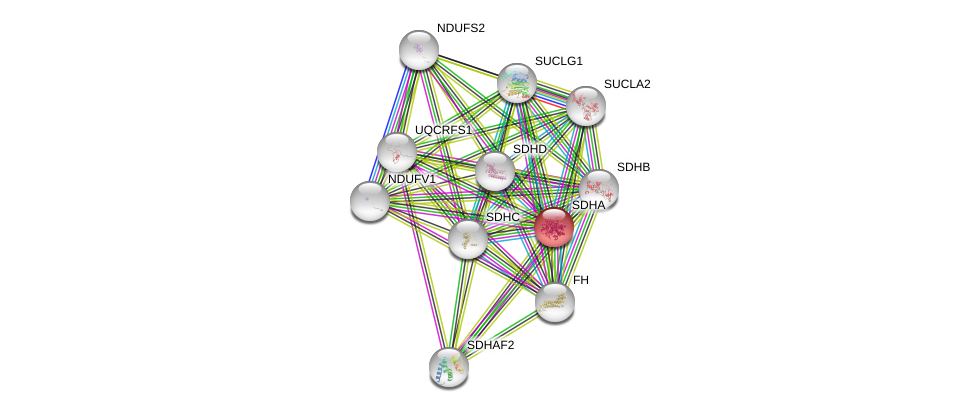
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000073578 | SDHA | 93 | 77.311 | WBGene00015391 | sdha-1 | 96 | 73.034 | Caenorhabditis_elegans |
| ENSG00000073578 | SDHA | 100 | 76.879 | FBgn0261439 | SdhA | 100 | 73.874 | Drosophila_melanogaster |
| ENSG00000073578 | SDHA | 100 | 96.701 | ENSMUSG00000021577 | Sdha | 99 | 96.667 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000104 | succinate dehydrogenase activity | 21873635. | IBA | Function |
| GO:0000104 | succinate dehydrogenase activity | 7550341. | IMP | Function |
| GO:0005515 | protein binding | 15961414.19628817.19688755.23747254.24606901.26496610.26749241.28330616. | IPI | Function |
| GO:0005730 | nucleolus | - | IDA | Component |
| GO:0005739 | mitochondrion | 7550341.16826196. | IDA | Component |
| GO:0005743 | mitochondrial inner membrane | - | ISS | Component |
| GO:0005743 | mitochondrial inner membrane | - | TAS | Component |
| GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | 21873635. | IBA | Component |
| GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | - | ISS | Component |
| GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | 7550341. | TAS | Component |
| GO:0006099 | tricarboxylic acid cycle | 21873635. | IBA | Process |
| GO:0006099 | tricarboxylic acid cycle | - | IEA | Process |
| GO:0006099 | tricarboxylic acid cycle | 7550341. | TAS | Process |
| GO:0006105 | succinate metabolic process | 7550341. | IDA | Process |
| GO:0006121 | mitochondrial electron transport, succinate to ubiquinone | 21873635. | IBA | Process |
| GO:0007399 | nervous system development | 16361598. | IMP | Process |
| GO:0008177 | succinate dehydrogenase (ubiquinone) activity | 21873635. | IBA | Function |
| GO:0008177 | succinate dehydrogenase (ubiquinone) activity | 24781757. | IMP | Function |
| GO:0009055 | electron transfer activity | 21873635. | IBA | Function |
| GO:0022904 | respiratory electron transport chain | 7550341. | IDA | Process |
| GO:0045282 | plasma membrane succinate dehydrogenase complex | 21873635. | IBA | Component |
| GO:0050660 | flavin adenine dinucleotide binding | 21873635. | IBA | Function |
| GO:0055114 | oxidation-reduction process | 7550341. | IDA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| BRCA | |||
| CESC | |||
| ESCA | |||
| HNSC | |||
| KIRP | |||
| LGG | |||
| LUSC | |||
| OV | |||
| SARC | |||
| SKCM | |||
| STAD | |||
| TGCT | |||
| THCA | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| ACC | ||
| HNSC | ||
| BRCA | ||
| KIRP | ||
| LAML | ||
| KICH | ||
| LGG | ||
| LUAD | ||
| UVM |