
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000557562 | FERMT2-214 | 555 | - | ENSP00000451085 | 65 (aa) | - | G3V379 |
| ENST00000343279 | FERMT2-202 | 3247 | XM_006720008 | ENSP00000342858 | 687 (aa) | XP_006720071 | Q96AC1 |
| ENST00000395631 | FERMT2-203 | 3369 | - | ENSP00000378993 | 680 (aa) | - | Q96AC1 |
| ENST00000553663 | FERMT2-206 | 655 | - | ENSP00000451134 | 133 (aa) | - | H0YJB6 |
| ENST00000557255 | FERMT2-213 | 6001 | - | - | - (aa) | - | - |
| ENST00000341590 | FERMT2-201 | 3338 | XM_005267285 | ENSP00000340391 | 680 (aa) | XP_005267342 | Q96AC1 |
| ENST00000553768 | FERMT2-207 | 791 | - | - | - (aa) | - | - |
| ENST00000555546 | FERMT2-211 | 2384 | - | - | - (aa) | - | - |
| ENST00000554288 | FERMT2-209 | 700 | - | ENSP00000451268 | 155 (aa) | - | G3V3J0 |
| ENST00000553373 | FERMT2-205 | 2064 | - | ENSP00000451084 | 687 (aa) | - | Q96AC1 |
| ENST00000554712 | FERMT2-210 | 587 | - | ENSP00000450506 | 153 (aa) | - | G3V281 |
| ENST00000554152 | FERMT2-208 | 3029 | - | ENSP00000450741 | 640 (aa) | - | H0YJ34 |
| ENST00000399304 | FERMT2-204 | 2082 | - | ENSP00000382243 | 633 (aa) | - | Q96AC1 |
| ENST00000555692 | FERMT2-212 | 754 | - | ENSP00000452472 | 133 (aa) | - | G3V5R2 |
| ENST00000635305 | FERMT2-215 | 2482 | - | ENSP00000489609 | 545 (aa) | - | A0A0U1RRM8 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000073712 | Blood Pressure | 3.4538510E-005 | - |
| ENSG00000073712 | Blood Pressure | 1.5866215E-005 | - |
| ENSG00000073712 | Blood Pressure | 4.9068987E-005 | - |
| ENSG00000073712 | Blood Pressure | 2.8678730E-005 | - |
| ENSG00000073712 | Blood Pressure | 3.7978313E-006 | - |
| ENSG00000073712 | Blood Pressure | 3.1925698E-006 | - |
| ENSG00000073712 | Blood Pressure | 3.2355810E-005 | - |
| ENSG00000073712 | Blood Pressure | 3.0310679E-006 | - |
| ENSG00000073712 | Prostatic Neoplasms | 2E-14 | 23535732 |
| ENSG00000073712 | Bronchodilator Agents | 5E-6 | 26634245 |
| ENSG00000073712 | Lung Volume Measurements | 5E-6 | 26634245 |
| ENSG00000073712 | Glaucoma, Angle-Closure | 3E-11 | 27064256 |
| ENSG00000073712 | Alzheimer Disease | 8E-9 | 24162737 |
| ENSG00000073712 | Glaucoma, Angle-Closure | 5E-9 | 27064256 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000073712 | rs8009633 | 14 | 52920118 | G | Intraocular pressure | 29617998 | [0.085-0.15] unit increase | 0.1173 | EFO_0004695 |
| ENSG00000073712 | rs79534246 | 14 | 52919724 | A | Post bronchodilator FEV1/FVC ratio | 26634245 | unit increase | 0.17 | GO_0097366|EFO_0004713 |
| ENSG00000073712 | rs7494379 | 14 | 52944673 | G | Glaucoma (primary angle closure) | 27064256 | [NR] | 1.14 | EFO_1001506 |
| ENSG00000073712 | rs7494379 | 14 | 52944673 | G | Glaucoma (primary angle closure) | 27064256 | [NR] | 1.13 | EFO_1001506 |
| ENSG00000073712 | rs8008270 | 14 | 52905612 | G | Prostate cancer | 23535732 | [1.08-1.16] | 1.12 | EFO_0001663 |
| ENSG00000073712 | rs17125944 | 14 | 52933911 | C | Alzheimer's disease (late onset) | 24162737 | [1.09-1.19] | 1.14 | EFO_0000249 |
| ENSG00000073712 | rs12147852 | 14 | 52895157 | ? | Intraocular pressure | 30054594 | [0.091-0.151] unit decrease | 0.1211 | EFO_0004695 |
| ENSG00000073712 | rs17125924 | 14 | 52924962 | ? | Alzheimer's disease or family history of Alzheimer's disease | 29777097 | EFO_0000249|EFO_0009268 | ||
| ENSG00000073712 | rs4901308 | 14 | 52913207 | T | PR interval | 30046033 | [0.51-1.09] ms decrease | 0.8 | EFO_0004462 |
| ENSG00000073712 | rs9888615 | 14 | 52910822 | T | Systolic blood pressure | 28135244 | [0.22-0.42] unit decrease | 0.318 | EFO_0006335 |
| ENSG00000073712 | rs8009633 | 14 | 52920118 | G | Intraocular pressure | 29785010 | [0.081-0.139] unit increase | 0.11 | EFO_0004695 |
| ENSG00000073712 | rs12887631 | 14 | 52906681 | ? | Systolic blood pressure | 30595370 | EFO_0006335 | ||
| ENSG00000073712 | rs7161323 | 14 | 52899431 | C | Diastolic blood pressure | 27841878 | unit decrease | 0.147 | EFO_0006336 |
| ENSG00000073712 | rs7161323 | 14 | 52899431 | C | Systolic blood pressure | 27841878 | unit decrease | 0.275 | EFO_0006335 |
| ENSG00000073712 | rs7161323 | 14 | 52899431 | C | Pulse pressure | 27841878 | unit decrease | 0.132 | EFO_0005763 |
| ENSG00000073712 | rs8008270 | 14 | 52905612 | C | Prostate cancer | 29892016 | [1.07-1.11] | 1.09 | EFO_0001663 |
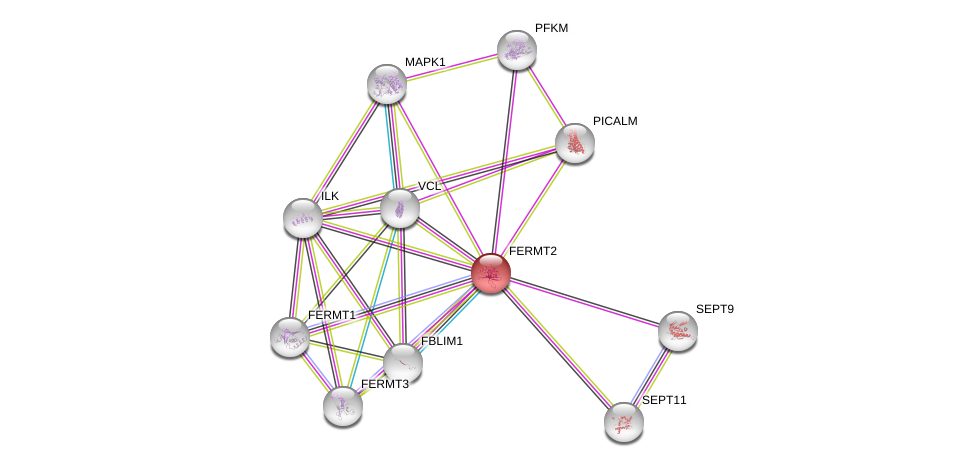
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001725 | stress fiber | - | IEA | Component |
| GO:0005515 | protein binding | 22699938. | IPI | Function |
| GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | 21325030.22030399. | IDA | Function |
| GO:0005634 | nucleus | 22699938. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | 22699938. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0005925 | focal adhesion | 21325030.29162887. | IDA | Component |
| GO:0005938 | cell cortex | - | IEA | Component |
| GO:0007160 | cell-matrix adhesion | 21325030. | IMP | Process |
| GO:0007160 | cell-matrix adhesion | - | ISS | Process |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 21325030. | IMP | Process |
| GO:0007229 | integrin-mediated signaling pathway | 21325030. | IMP | Process |
| GO:0008360 | regulation of cell shape | - | IEA | Process |
| GO:0009986 | cell surface | - | IEA | Component |
| GO:0016055 | Wnt signaling pathway | 22699938. | IMP | Process |
| GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | 21325030. | IDA | Component |
| GO:0031258 | lamellipodium membrane | - | IEA | Component |
| GO:0031674 | I band | - | IEA | Component |
| GO:0033622 | integrin activation | 22030399. | IMP | Process |
| GO:0034329 | cell junction assembly | - | TAS | Process |
| GO:0034446 | substrate adhesion-dependent cell spreading | - | ISS | Process |
| GO:0048041 | focal adhesion assembly | - | ISS | Process |
| GO:0051015 | actin filament binding | - | IEA | Function |
| GO:0072657 | protein localization to membrane | - | ISS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| KIRP | |||
| MESO | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| KIRP | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LGG |