
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000617585 | ARHGEF1-223 | 737 | - | - | - (aa) | - | - |
| ENST00000598587 | ARHGEF1-216 | 359 | - | - | - (aa) | - | - |
| ENST00000599846 | ARHGEF1-219 | 3393 | - | ENSP00000470715 | 968 (aa) | - | M0QZR4 |
| ENST00000594258 | ARHGEF1-207 | 919 | - | ENSP00000470042 | 307 (aa) | - | M0QYS3 |
| ENST00000347545 | ARHGEF1-202 | 3091 | - | ENSP00000344429 | 879 (aa) | - | Q92888 |
| ENST00000598444 | ARHGEF1-215 | 425 | - | - | - (aa) | - | - |
| ENST00000594513 | ARHGEF1-209 | 325 | - | - | - (aa) | - | - |
| ENST00000595897 | ARHGEF1-213 | 2973 | - | - | - (aa) | - | - |
| ENST00000354532 | ARHGEF1-203 | 3243 | - | ENSP00000346532 | 912 (aa) | - | Q92888 |
| ENST00000596957 | ARHGEF1-214 | 570 | - | - | - (aa) | - | - |
| ENST00000599589 | ARHGEF1-218 | 2636 | - | ENSP00000469735 | 702 (aa) | - | M0QYC1 |
| ENST00000593609 | ARHGEF1-205 | 576 | - | - | - (aa) | - | - |
| ENST00000594044 | ARHGEF1-206 | 834 | - | - | - (aa) | - | - |
| ENST00000600387 | ARHGEF1-221 | 2272 | - | - | - (aa) | - | - |
| ENST00000598812 | ARHGEF1-217 | 588 | - | ENSP00000473229 | 88 (aa) | - | M0R3H5 |
| ENST00000378152 | ARHGEF1-204 | 2986 | - | ENSP00000367394 | 948 (aa) | - | Q92888 |
| ENST00000594417 | ARHGEF1-208 | 769 | - | ENSP00000471942 | 128 (aa) | - | M0R1K8 |
| ENST00000337665 | ARHGEF1-201 | 3171 | - | ENSP00000337261 | 927 (aa) | - | Q92888 |
| ENST00000594521 | ARHGEF1-210 | 826 | - | ENSP00000470557 | 275 (aa) | - | M0QZH8 |
| ENST00000595723 | ARHGEF1-211 | 872 | - | ENSP00000472466 | 291 (aa) | - | M0R2C7 |
| ENST00000595770 | ARHGEF1-212 | 663 | NR_135626 | - | - (aa) | - | - |
| ENST00000600274 | ARHGEF1-220 | 3439 | - | - | - (aa) | - | - |
| ENST00000600517 | ARHGEF1-222 | 428 | - | ENSP00000469409 | 68 (aa) | - | M0QXV6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04270 | Vascular smooth muscle contraction | KEGG |
| hsa04611 | Platelet activation | KEGG |
| hsa04810 | Regulation of actin cytoskeleton | KEGG |
| hsa04928 | Parathyroid hormone synthesis, secretion and action | KEGG |
| hsa05163 | Human cytomegalovirus infection | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| hsa05205 | Proteoglycans in cancer | KEGG |
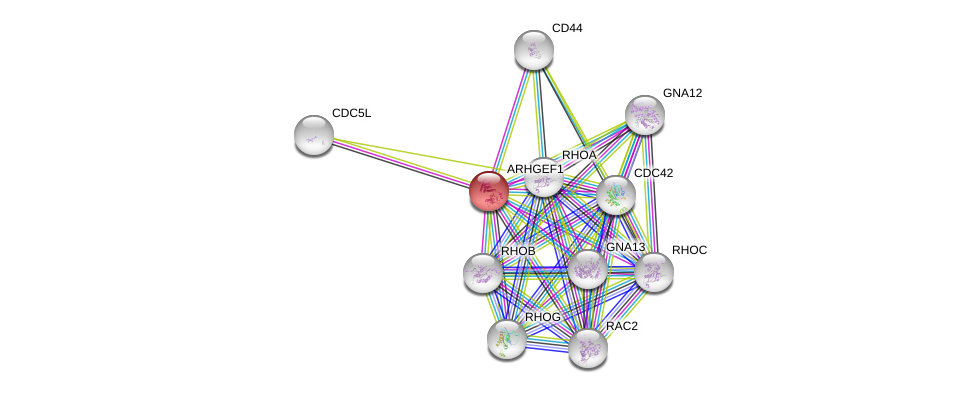
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000076928 | ARHGEF1 | 99 | 89.364 | ENSMUSG00000040940 | Arhgef1 | 98 | 91.304 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001664 | G protein-coupled receptor binding | - | IEA | Function |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | - | IEA | Function |
| GO:0005096 | GTPase activator activity | - | IEA | Function |
| GO:0005515 | protein binding | 12681510.20300064.25416956. | IPI | Function |
| GO:0005737 | cytoplasm | 10747909. | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0007186 | G protein-coupled receptor signaling pathway | - | TAS | Process |
| GO:0007266 | Rho protein signal transduction | 10747909. | TAS | Process |
| GO:0008283 | cell proliferation | 8810315. | TAS | Process |
| GO:0035023 | regulation of Rho protein signal transduction | - | IEA | Process |
| GO:0043065 | positive regulation of apoptotic process | - | TAS | Process |
| GO:0043547 | positive regulation of GTPase activity | - | IEA | Process |
| GO:0051056 | regulation of small GTPase mediated signal transduction | - | TAS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| READ | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| LUSC | ||
| BRCA | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| OV |