
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000483329 | ResIII | PF04851.15 | 0.00022 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 21673954 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000411466 | SRCAP-203 | 836 | - | ENSP00000405186 | 100 (aa) | - | C9J4U4 |
| ENST00000395059 | SRCAP-202 | 9126 | - | ENSP00000378499 | 2971 (aa) | - | A0A0A0MS59 |
| ENST00000262518 | SRCAP-201 | 11754 | - | ENSP00000262518 | 3230 (aa) | - | Q6ZRS2 |
| ENST00000474008 | SRCAP-204 | 730 | - | - | - (aa) | - | - |
| ENST00000483083 | SRCAP-205 | 5937 | - | ENSP00000483329 | 1610 (aa) | - | A0A087X0E3 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000080603 | rs8058578 | 16 | 30714927 | A | Systemic lupus erythematosus | 28714469 | [1.06-1.16] | 1.11 | EFO_0002690 |
| ENSG00000080603 | rs8058578 | 16 | 30714927 | T | Multiple myeloma | 27363682 | [NR] | 1.1 | EFO_0001378 |
| ENSG00000080603 | rs113715277 | 16 | 30701667 | T | Pulse pressure | 30578418 | [0.14-0.26] mmHg increase | 0.2026 | EFO_0005763 |
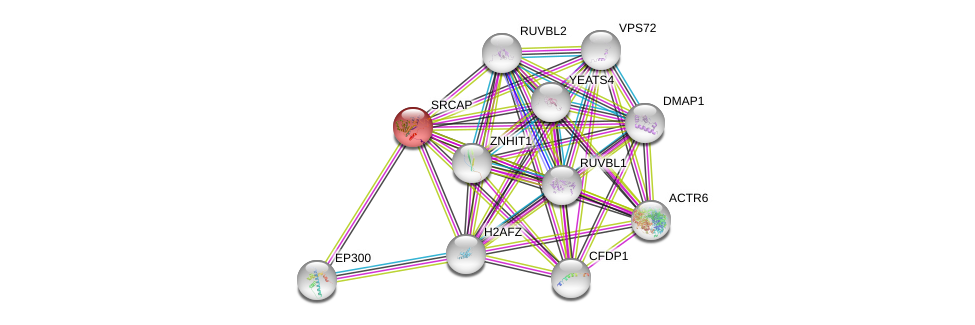
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000080603 | SRCAP | 59 | 93.220 | ENSSTOG00000020765 | - | 99 | 90.074 | Ictidomys_tridecemlineatus |
| ENSG00000080603 | SRCAP | 100 | 90.000 | MGP_CAROLIEiJ_G0030507 | - | 100 | 88.712 | Mus_caroli |
| ENSG00000080603 | SRCAP | 100 | 71.000 | ENSNGAG00000022162 | - | 99 | 85.177 | Nannospalax_galili |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000812 | Swr1 complex | 21873635. | IBA | Component |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0003713 | transcription coactivator activity | 10347196. | TAS | Function |
| GO:0004386 | helicase activity | - | IEA | Function |
| GO:0004402 | histone acetyltransferase activity | 10347196. | TAS | Function |
| GO:0005515 | protein binding | 10702287. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 10702287. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005794 | Golgi apparatus | - | IDA | Component |
| GO:0006338 | chromatin remodeling | 21873635. | IBA | Process |
| GO:0006357 | regulation of transcription by RNA polymerase II | 10347196. | TAS | Process |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016458 | gene silencing | 21873635. | IBA | Process |
| GO:0016573 | histone acetylation | - | IEA | Process |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0016887 | ATPase activity | 21873635. | IBA | Function |
| GO:0032991 | protein-containing complex | 14966270. | IDA | Component |
| GO:0042393 | histone binding | 21873635. | IBA | Function |
| GO:0043044 | ATP-dependent chromatin remodeling | 21873635. | IBA | Process |
| GO:0043486 | histone exchange | 21873635. | IBA | Process |
| GO:0048471 | perinuclear region of cytoplasm | 10702287. | IDA | Component |
| GO:1903508 | positive regulation of nucleic acid-templated transcription | - | IEA | Process |