
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000354877 | RIO1 | PF01163.22 | 0.00011 | 1 | 1 |
| ENSP00000378914 | RIO1 | PF01163.22 | 0.00011 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 30917721 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000574732 | ULK2-207 | 564 | - | ENSP00000465666 | 21 (aa) | - | K7EKK9 |
| ENST00000574854 | ULK2-208 | 493 | - | - | - (aa) | - | - |
| ENST00000575432 | ULK2-209 | 594 | - | ENSP00000464892 | 198 (aa) | - | K7EIU2 |
| ENST00000395544 | ULK2-202 | 6010 | XM_011524087 | ENSP00000378914 | 1036 (aa) | XP_011522389 | Q8IYT8 |
| ENST00000571137 | ULK2-204 | 615 | - | - | - (aa) | - | - |
| ENST00000580118 | ULK2-210 | 577 | - | ENSP00000467277 | 49 (aa) | - | K7EP89 |
| ENST00000571177 | ULK2-205 | 492 | - | - | - (aa) | - | - |
| ENST00000580130 | ULK2-211 | 627 | - | - | - (aa) | - | - |
| ENST00000570983 | ULK2-203 | 3096 | - | - | - (aa) | - | - |
| ENST00000361658 | ULK2-201 | 3908 | XM_017025425 | ENSP00000354877 | 1036 (aa) | XP_016880914 | Q8IYT8 |
| ENST00000571454 | ULK2-206 | 562 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000083290 | Parkinson Disease | 2.505542E-5 | 17052657 |
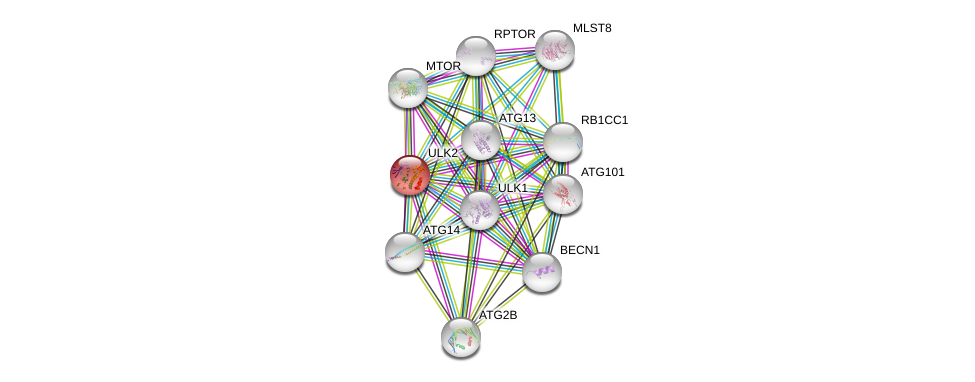
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000083290 | ULK2 | 100 | 72.993 | ENSAPOG00000004920 | ulk2 | 99 | 73.491 | Acanthochromis_polyacanthus |
| ENSG00000083290 | ULK2 | 97 | 93.545 | ENSAMEG00000004430 | ULK2 | 100 | 93.545 | Ailuropoda_melanoleuca |
| ENSG00000083290 | ULK2 | 100 | 73.088 | ENSAOCG00000006514 | ulk2 | 99 | 73.585 | Amphiprion_ocellaris |
| ENSG00000083290 | ULK2 | 97 | 86.871 | ENSAPLG00000003309 | ULK2 | 99 | 86.871 | Anas_platyrhynchos |
| ENSG00000083290 | ULK2 | 100 | 62.946 | ENSACAG00000000631 | ULK2 | 87 | 93.609 | Anolis_carolinensis |
| ENSG00000083290 | ULK2 | 100 | 72.590 | ENSACLG00000020477 | ulk2 | 99 | 72.805 | Astatotilapia_calliptera |
| ENSG00000083290 | ULK2 | 100 | 74.525 | ENSAMXG00000009215 | ulk2 | 99 | 74.905 | Astyanax_mexicanus |
| ENSG00000083290 | ULK2 | 100 | 96.622 | ENSCJAG00000011832 | ULK2 | 100 | 96.622 | Callithrix_jacchus |
| ENSG00000083290 | ULK2 | 100 | 94.793 | ENSCAFG00000018126 | ULK2 | 100 | 94.793 | Canis_familiaris |
| ENSG00000083290 | ULK2 | 100 | 89.479 | ENSCHIG00000011283 | ULK2 | 100 | 89.479 | Capra_hircus |
| ENSG00000083290 | ULK2 | 99 | 95.141 | ENSTSYG00000003217 | ULK2 | 99 | 95.044 | Carlito_syrichta |
| ENSG00000083290 | ULK2 | 100 | 93.147 | ENSCPOG00000010048 | ULK2 | 100 | 93.147 | Cavia_porcellus |
| ENSG00000083290 | ULK2 | 97 | 96.875 | ENSCATG00000032753 | ULK2 | 100 | 96.875 | Cercocebus_atys |
| ENSG00000083290 | ULK2 | 97 | 93.539 | ENSCLAG00000008483 | ULK2 | 100 | 93.539 | Chinchilla_lanigera |
| ENSG00000083290 | ULK2 | 100 | 97.973 | ENSCSAG00000007819 | ULK2 | 100 | 97.973 | Chlorocebus_sabaeus |
| ENSG00000083290 | ULK2 | 83 | 90.647 | ENSCHOG00000012625 | ULK2 | 85 | 87.050 | Choloepus_hoffmanni |
| ENSG00000083290 | ULK2 | 100 | 84.716 | ENSCPBG00000015808 | ULK2 | 99 | 84.716 | Chrysemys_picta_bellii |
| ENSG00000083290 | ULK2 | 100 | 94.208 | ENSCGRG00001011469 | Ulk2 | 100 | 94.208 | Cricetulus_griseus_chok1gshd |
| ENSG00000083290 | ULK2 | 97 | 94.036 | ENSCGRG00000009104 | Ulk2 | 100 | 94.036 | Cricetulus_griseus_crigri |
| ENSG00000083290 | ULK2 | 100 | 73.926 | ENSDARG00000097205 | ulk2 | 99 | 74.141 | Danio_rerio |
| ENSG00000083290 | ULK2 | 97 | 91.576 | ENSDNOG00000048497 | ULK2 | 61 | 91.576 | Dasypus_novemcinctus |
| ENSG00000083290 | ULK2 | 100 | 93.919 | ENSDORG00000012863 | Ulk2 | 100 | 93.919 | Dipodomys_ordii |
| ENSG00000083290 | ULK2 | 84 | 89.103 | ENSETEG00000002660 | ULK2 | 86 | 95.440 | Echinops_telfairi |
| ENSG00000083290 | ULK2 | 100 | 94.788 | ENSEASG00005017583 | ULK2 | 100 | 94.788 | Equus_asinus_asinus |
| ENSG00000083290 | ULK2 | 100 | 94.691 | ENSECAG00000008882 | ULK2 | 100 | 92.687 | Equus_caballus |
| ENSG00000083290 | ULK2 | 100 | 73.764 | ENSELUG00000017192 | ulk2 | 99 | 73.979 | Esox_lucius |
| ENSG00000083290 | ULK2 | 100 | 93.173 | ENSFCAG00000005190 | ULK2 | 100 | 93.173 | Felis_catus |
| ENSG00000083290 | ULK2 | 100 | 83.784 | ENSFALG00000005884 | ULK2 | 99 | 83.784 | Ficedula_albicollis |
| ENSG00000083290 | ULK2 | 100 | 93.822 | ENSFDAG00000004531 | ULK2 | 98 | 93.822 | Fukomys_damarensis |
| ENSG00000083290 | ULK2 | 100 | 71.321 | ENSFHEG00000001152 | ulk2 | 99 | 71.631 | Fundulus_heteroclitus |
| ENSG00000083290 | ULK2 | 100 | 72.060 | ENSGMOG00000002637 | ulk2 | 100 | 72.274 | Gadus_morhua |
| ENSG00000083290 | ULK2 | 99 | 71.851 | ENSGAFG00000019721 | ulk2 | 98 | 72.069 | Gambusia_affinis |
| ENSG00000083290 | ULK2 | 100 | 72.049 | ENSGACG00000013524 | ulk2 | 100 | 72.170 | Gasterosteus_aculeatus |
| ENSG00000083290 | ULK2 | 100 | 84.623 | ENSGAGG00000011573 | ULK2 | 99 | 84.623 | Gopherus_agassizii |
| ENSG00000083290 | ULK2 | 100 | 99.614 | ENSGGOG00000007689 | ULK2 | 100 | 99.614 | Gorilla_gorilla |
| ENSG00000083290 | ULK2 | 100 | 72.495 | ENSHBUG00000006775 | ulk2 | 99 | 72.710 | Haplochromis_burtoni |
| ENSG00000083290 | ULK2 | 100 | 73.594 | ENSIPUG00000007930 | ulk2 | 99 | 74.000 | Ictalurus_punctatus |
| ENSG00000083290 | ULK2 | 100 | 94.981 | ENSSTOG00000007174 | ULK2 | 100 | 94.981 | Ictidomys_tridecemlineatus |
| ENSG00000083290 | ULK2 | 100 | 72.212 | ENSKMAG00000002479 | ulk2 | 99 | 72.427 | Kryptolebias_marmoratus |
| ENSG00000083290 | ULK2 | 100 | 79.048 | ENSLACG00000001863 | ULK2 | 99 | 79.143 | Latimeria_chalumnae |
| ENSG00000083290 | ULK2 | 100 | 74.597 | ENSLOCG00000005372 | ulk2 | 99 | 75.000 | Lepisosteus_oculatus |
| ENSG00000083290 | ULK2 | 100 | 98.069 | ENSMFAG00000037593 | ULK2 | 100 | 98.069 | Macaca_fascicularis |
| ENSG00000083290 | ULK2 | 100 | 98.069 | ENSMMUG00000021156 | ULK2 | 100 | 98.069 | Macaca_mulatta |
| ENSG00000083290 | ULK2 | 100 | 97.394 | ENSMNEG00000030857 | ULK2 | 100 | 97.394 | Macaca_nemestrina |
| ENSG00000083290 | ULK2 | 97 | 98.012 | ENSMLEG00000041408 | ULK2 | 100 | 98.012 | Mandrillus_leucophaeus |
| ENSG00000083290 | ULK2 | 100 | 72.091 | ENSMAMG00000002940 | ulk2 | 99 | 72.753 | Mastacembelus_armatus |
| ENSG00000083290 | ULK2 | 100 | 72.590 | ENSMZEG00005001956 | ulk2 | 99 | 72.805 | Maylandia_zebra |
| ENSG00000083290 | ULK2 | 100 | 86.481 | ENSMGAG00000006805 | ULK2 | 100 | 86.481 | Meleagris_gallopavo |
| ENSG00000083290 | ULK2 | 94 | 94.964 | ENSMICG00000031631 | ULK2 | 99 | 94.964 | Microcebus_murinus |
| ENSG00000083290 | ULK2 | 100 | 93.726 | ENSMOCG00000012863 | Ulk2 | 100 | 93.726 | Microtus_ochrogaster |
| ENSG00000083290 | ULK2 | 100 | 89.317 | ENSMODG00000010934 | ULK2 | 99 | 89.317 | Monodelphis_domestica |
| ENSG00000083290 | ULK2 | 100 | 72.375 | ENSMALG00000019136 | ulk2 | 99 | 72.968 | Monopterus_albus |
| ENSG00000083290 | ULK2 | 100 | 93.443 | MGP_CAROLIEiJ_G0016485 | Ulk2 | 100 | 93.443 | Mus_caroli |
| ENSG00000083290 | ULK2 | 100 | 93.635 | ENSMUSG00000004798 | Ulk2 | 100 | 93.635 | Mus_musculus |
| ENSG00000083290 | ULK2 | 100 | 93.539 | MGP_SPRETEiJ_G0017320 | Ulk2 | 100 | 93.539 | Mus_spretus |
| ENSG00000083290 | ULK2 | 94 | 83.750 | ENSMLUG00000007422 | - | 100 | 83.750 | Myotis_lucifugus |
| ENSG00000083290 | ULK2 | 97 | 89.812 | ENSMLUG00000017762 | - | 99 | 89.812 | Myotis_lucifugus |
| ENSG00000083290 | ULK2 | 100 | 94.305 | ENSNGAG00000009072 | Ulk2 | 100 | 94.305 | Nannospalax_galili |
| ENSG00000083290 | ULK2 | 100 | 93.629 | ENSODEG00000000757 | ULK2 | 100 | 93.629 | Octodon_degus |
| ENSG00000083290 | ULK2 | 100 | 72.495 | ENSONIG00000010482 | ulk2 | 99 | 72.710 | Oreochromis_niloticus |
| ENSG00000083290 | ULK2 | 100 | 94.884 | ENSOCUG00000013388 | ULK2 | 100 | 95.174 | Oryctolagus_cuniculus |
| ENSG00000083290 | ULK2 | 100 | 70.833 | ENSORLG00000014987 | ulk2 | 99 | 71.117 | Oryzias_latipes |
| ENSG00000083290 | ULK2 | 100 | 70.644 | ENSORLG00020008437 | ulk2 | 99 | 70.928 | Oryzias_latipes_hni |
| ENSG00000083290 | ULK2 | 100 | 70.739 | ENSORLG00015014264 | ulk2 | 99 | 71.023 | Oryzias_latipes_hsok |
| ENSG00000083290 | ULK2 | 100 | 71.429 | ENSOMEG00000008411 | ulk2 | 99 | 71.429 | Oryzias_melastigma |
| ENSG00000083290 | ULK2 | 99 | 92.967 | ENSOGAG00000017081 | ULK2 | 99 | 92.967 | Otolemur_garnettii |
| ENSG00000083290 | ULK2 | 97 | 88.340 | ENSOARG00000017770 | ULK2 | 100 | 88.340 | Ovis_aries |
| ENSG00000083290 | ULK2 | 97 | 99.503 | ENSPPAG00000037047 | ULK2 | 100 | 99.503 | Pan_paniscus |
| ENSG00000083290 | ULK2 | 97 | 93.168 | ENSPTIG00000012203 | ULK2 | 97 | 93.168 | Panthera_tigris_altaica |
| ENSG00000083290 | ULK2 | 100 | 99.517 | ENSPTRG00000008875 | ULK2 | 100 | 99.517 | Pan_troglodytes |
| ENSG00000083290 | ULK2 | 100 | 98.166 | ENSPANG00000023620 | ULK2 | 100 | 98.166 | Papio_anubis |
| ENSG00000083290 | ULK2 | 92 | 65.106 | ENSPKIG00000018902 | - | 56 | 65.106 | Paramormyrops_kingsleyae |
| ENSG00000083290 | ULK2 | 97 | 86.673 | ENSPSIG00000015566 | ULK2 | 99 | 86.772 | Pelodiscus_sinensis |
| ENSG00000083290 | ULK2 | 100 | 93.070 | ENSPEMG00000016139 | Ulk2 | 100 | 93.070 | Peromyscus_maniculatus_bairdii |
| ENSG00000083290 | ULK2 | 100 | 89.028 | ENSPCIG00000014203 | ULK2 | 99 | 89.028 | Phascolarctos_cinereus |
| ENSG00000083290 | ULK2 | 100 | 71.226 | ENSPFOG00000016656 | ulk2 | 99 | 71.536 | Poecilia_formosa |
| ENSG00000083290 | ULK2 | 100 | 71.604 | ENSPREG00000015581 | ulk2 | 99 | 71.913 | Poecilia_reticulata |
| ENSG00000083290 | ULK2 | 100 | 94.691 | ENSPCOG00000014882 | ULK2 | 100 | 94.691 | Propithecus_coquereli |
| ENSG00000083290 | ULK2 | 100 | 72.495 | ENSPNYG00000023925 | ulk2 | 99 | 72.710 | Pundamilia_nyererei |
| ENSG00000083290 | ULK2 | 100 | 75.143 | ENSPNAG00000007165 | ulk2 | 99 | 75.357 | Pygocentrus_nattereri |
| ENSG00000083290 | ULK2 | 100 | 93.828 | ENSRNOG00000002763 | Ulk2 | 100 | 93.828 | Rattus_norvegicus |
| ENSG00000083290 | ULK2 | 97 | 89.219 | ENSSHAG00000003186 | ULK2 | 100 | 89.219 | Sarcophilus_harrisii |
| ENSG00000083290 | ULK2 | 100 | 72.144 | ENSSMAG00000013811 | ulk2 | 100 | 73.007 | Scophthalmus_maximus |
| ENSG00000083290 | ULK2 | 100 | 72.710 | ENSSLDG00000011160 | ulk2 | 99 | 72.925 | Seriola_lalandi_dorsalis |
| ENSG00000083290 | ULK2 | 100 | 72.049 | ENSSPAG00000010229 | ulk2 | 99 | 72.427 | Stegastes_partitus |
| ENSG00000083290 | ULK2 | 100 | 84.119 | ENSSSCG00000018045 | ULK2 | 100 | 83.542 | Sus_scrofa |
| ENSG00000083290 | ULK2 | 99 | 86.403 | ENSTGUG00000007111 | - | 99 | 86.403 | Taeniopygia_guttata |
| ENSG00000083290 | ULK2 | 99 | 68.994 | ENSTNIG00000014387 | ulk2 | 99 | 69.367 | Tetraodon_nigroviridis |
| ENSG00000083290 | ULK2 | 78 | 82.759 | ENSVPAG00000007776 | - | 84 | 73.699 | Vicugna_pacos |
| ENSG00000083290 | ULK2 | 97 | 94.362 | ENSVVUG00000030016 | ULK2 | 93 | 94.362 | Vulpes_vulpes |
| ENSG00000083290 | ULK2 | 100 | 78.011 | ENSXETG00000032595 | ULK2 | 100 | 78.298 | Xenopus_tropicalis |
| ENSG00000083290 | ULK2 | 100 | 72.196 | ENSXMAG00000005775 | ulk2 | 99 | 72.411 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000045 | autophagosome assembly | 21873635. | IBA | Process |
| GO:0000407 | phagophore assembly site | 21873635. | IBA | Component |
| GO:0004674 | protein serine/threonine kinase activity | 21873635. | IBA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 18936157. | IDA | Function |
| GO:0005515 | protein binding | 18936157. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0006914 | autophagy | 18936157. | IDA | Process |
| GO:0007165 | signal transduction | - | IEA | Process |
| GO:0010506 | regulation of autophagy | 21873635. | IBA | Process |
| GO:0016020 | membrane | 21873635. | IBA | Component |
| GO:0030659 | cytoplasmic vesicle membrane | - | IEA | Component |
| GO:0034045 | phagophore assembly site membrane | 18936157. | IDA | Component |
| GO:0042594 | response to starvation | - | ISS | Process |
| GO:0046777 | protein autophosphorylation | 21873635. | IBA | Process |
| GO:0046777 | protein autophosphorylation | 18936157. | IDA | Process |
| GO:0048671 | negative regulation of collateral sprouting | - | IEA | Process |
| GO:0048675 | axon extension | 21873635. | IBA | Process |
| GO:0075044 | autophagy of host cells involved in interaction with symbiont | 28389568. | IGI | Process |