
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000410630 | MMR_HSR1 | PF01926.23 | 4.3e-06 | 1 | 1 |
| ENSP00000364077 | MMR_HSR1 | PF01926.23 | 8.2e-06 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 31205503 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000414239 | RRAGB-203 | 644 | - | ENSP00000410630 | 194 (aa) | - | Q5VZM0 |
| ENST00000262850 | RRAGB-201 | 1573 | XM_017029218 | ENSP00000262850 | 374 (aa) | XP_016884707 | Q5VZM2 |
| ENST00000498762 | RRAGB-206 | 507 | - | - | - (aa) | - | - |
| ENST00000474757 | RRAGB-204 | 709 | - | - | - (aa) | - | - |
| ENST00000374941 | RRAGB-202 | 2061 | - | ENSP00000364077 | 346 (aa) | - | Q5VZM2 |
| ENST00000475061 | RRAGB-205 | 376 | - | - | - (aa) | - | - |
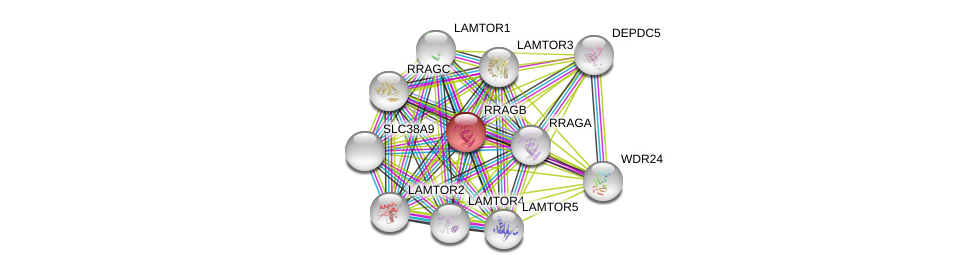
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSAPOG00000012140 | rraga | 99 | 95.527 | Acanthochromis_polyacanthus |
| ENSG00000083750 | RRAGB | 100 | 96.392 | ENSACIG00000013561 | rraga | 99 | 95.208 | Amphilophus_citrinellus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSAOCG00000002304 | rraga | 99 | 95.527 | Amphiprion_ocellaris |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSAPEG00000006571 | rraga | 99 | 95.527 | Amphiprion_percula |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSATEG00000011885 | rraga | 99 | 95.527 | Anabas_testudineus |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSANAG00000031746 | - | 100 | 97.125 | Aotus_nancymaae |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSANAG00000020912 | RRAGB | 100 | 99.422 | Aotus_nancymaae |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSACLG00000010458 | rraga | 99 | 95.847 | Astatotilapia_calliptera |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSAMXG00000030179 | rraga | 99 | 95.208 | Astyanax_mexicanus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSBTAG00000012980 | RRAGB | 100 | 99.133 | Bos_taurus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSBTAG00000033543 | RRAGA | 100 | 98.083 | Bos_taurus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSBTAG00000038064 | - | 100 | 97.764 | Bos_taurus |
| ENSG00000083750 | RRAGB | 99 | 67.157 | WBGene00006414 | raga-1 | 98 | 64.706 | Caenorhabditis_elegans |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSCJAG00000007317 | RRAGB | 100 | 99.133 | Callithrix_jacchus |
| ENSG00000083750 | RRAGB | 100 | 97.938 | ENSCJAG00000001550 | - | 100 | 96.805 | Callithrix_jacchus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCJAG00000022413 | - | 95 | 98.089 | Callithrix_jacchus |
| ENSG00000083750 | RRAGB | 100 | 99.485 | ENSCAFG00020013782 | RRAGB | 99 | 98.841 | Canis_lupus_dingo |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCAFG00020012925 | RRAGA | 100 | 98.083 | Canis_lupus_dingo |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCHIG00000004994 | RRAGA | 100 | 98.083 | Capra_hircus |
| ENSG00000083750 | RRAGB | 100 | 99.485 | ENSCHIG00000018786 | RRAGB | 100 | 98.844 | Capra_hircus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSTSYG00000004397 | RRAGB | 100 | 99.422 | Carlito_syrichta |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCPOG00000008047 | RRAGA | 100 | 98.083 | Cavia_porcellus |
| ENSG00000083750 | RRAGB | 98 | 96.316 | ENSCPOG00000011266 | RRAGB | 99 | 97.690 | Cavia_porcellus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSCCAG00000026742 | RRAGB | 99 | 99.130 | Cebus_capucinus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCCAG00000000619 | - | 100 | 98.083 | Cebus_capucinus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCATG00000015913 | RRAGA | 100 | 98.083 | Cercocebus_atys |
| ENSG00000083750 | RRAGB | 100 | 97.938 | ENSCLAG00000003913 | RRAGB | 100 | 97.399 | Chinchilla_lanigera |
| ENSG00000083750 | RRAGB | 100 | 97.938 | ENSCLAG00000017884 | RRAGA | 100 | 97.125 | Chinchilla_lanigera |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCHOG00000001957 | RRAGA | 100 | 98.083 | Choloepus_hoffmanni |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCPBG00000010507 | RRAGB | 100 | 97.764 | Chrysemys_picta_bellii |
| ENSG00000083750 | RRAGB | 99 | 86.010 | ENSCING00000007876 | - | 95 | 83.946 | Ciona_intestinalis |
| ENSG00000083750 | RRAGB | 99 | 85.492 | ENSCSAVG00000003505 | - | 96 | 83.278 | Ciona_savignyi |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSCANG00000017510 | RRAGB | 100 | 99.711 | Colobus_angolensis_palliatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCANG00000038134 | RRAGA | 100 | 98.083 | Colobus_angolensis_palliatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSCGRG00001013394 | Rragb | 99 | 99.342 | Cricetulus_griseus_chok1gshd |
| ENSG00000083750 | RRAGB | 98 | 98.947 | ENSCGRG00000006827 | Rragb | 99 | 99.342 | Cricetulus_griseus_crigri |
| ENSG00000083750 | RRAGB | 96 | 98.925 | ENSCGRG00000003443 | Rraga | 96 | 98.333 | Cricetulus_griseus_crigri |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSCSEG00000014142 | rraga | 99 | 95.847 | Cynoglossus_semilaevis |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSCVAG00000021719 | rraga | 99 | 95.208 | Cyprinodon_variegatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSDNOG00000015581 | RRAGA | 100 | 98.083 | Dasypus_novemcinctus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSDORG00000015994 | Rraga | 100 | 98.083 | Dipodomys_ordii |
| ENSG00000083750 | RRAGB | 99 | 98.964 | ENSDORG00000015932 | Rragb | 95 | 99.349 | Dipodomys_ordii |
| ENSG00000083750 | RRAGB | 100 | 79.381 | FBgn0037647 | RagA-B | 95 | 80.743 | Drosophila_melanogaster |
| ENSG00000083750 | RRAGB | 98 | 90.000 | ENSEBUG00000008941 | rraga | 89 | 87.346 | Eptatretus_burgeri |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSEASG00005019266 | RRAGA | 99 | 91.254 | Equus_asinus_asinus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSEASG00005006973 | RRAGB | 83 | 100.000 | Equus_asinus_asinus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSECAG00000005729 | RRAGA | 92 | 97.771 | Equus_caballus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSECAG00000006020 | RRAGB | 100 | 98.844 | Equus_caballus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSEEUG00000009466 | RRAGA | 100 | 98.083 | Erinaceus_europaeus |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSELUG00000009655 | rraga | 99 | 95.847 | Esox_lucius |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSFCAG00000022742 | RRAGA | 100 | 98.083 | Felis_catus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSFCAG00000013932 | RRAGB | 100 | 98.555 | Felis_catus |
| ENSG00000083750 | RRAGB | 100 | 98.454 | ENSFALG00000011263 | RRAGB | 87 | 96.865 | Ficedula_albicollis |
| ENSG00000083750 | RRAGB | 98 | 99.476 | ENSFDAG00000018157 | RRAGB | 99 | 99.672 | Fukomys_damarensis |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSFHEG00000002145 | rraga | 99 | 95.527 | Fundulus_heteroclitus |
| ENSG00000083750 | RRAGB | 100 | 96.392 | ENSGMOG00000004849 | rraga | 99 | 94.904 | Gadus_morhua |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSGALG00000041226 | RRAGB | 100 | 97.764 | Gallus_gallus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSGAFG00000006618 | rraga | 99 | 96.129 | Gambusia_affinis |
| ENSG00000083750 | RRAGB | 100 | 96.392 | ENSGACG00000020342 | rraga | 99 | 94.603 | Gasterosteus_aculeatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSGAGG00000012251 | RRAGB | 100 | 97.764 | Gopherus_agassizii |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSGGOG00000011501 | RRAGB | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSGGOG00000021937 | RRAGA | 100 | 98.083 | Gorilla_gorilla |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSHBUG00000004494 | rraga | 92 | 92.053 | Haplochromis_burtoni |
| ENSG00000083750 | RRAGB | 100 | 99.485 | ENSHGLG00000000313 | RRAGB | 100 | 98.266 | Heterocephalus_glaber_female |
| ENSG00000083750 | RRAGB | 100 | 98.454 | ENSHGLG00100020985 | RRAGA | 100 | 97.764 | Heterocephalus_glaber_male |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSHCOG00000006819 | rraga | 99 | 95.208 | Hippocampus_comes |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSIPUG00000014734 | Rraga | 99 | 95.527 | Ictalurus_punctatus |
| ENSG00000083750 | RRAGB | 96 | 100.000 | ENSSTOG00000004660 | RRAGB | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000083750 | RRAGB | 99 | 98.958 | ENSJJAG00000021386 | Rragb | 99 | 98.071 | Jaculus_jaculus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSJJAG00000008212 | Rraga | 100 | 98.083 | Jaculus_jaculus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSKMAG00000005709 | rraga | 99 | 95.527 | Kryptolebias_marmoratus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSLBEG00000004506 | rraga | 99 | 95.527 | Labrus_bergylta |
| ENSG00000083750 | RRAGB | 100 | 97.938 | ENSLACG00000016067 | RRAGB | 100 | 96.805 | Latimeria_chalumnae |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSLOCG00000014979 | rraga | 99 | 95.847 | Lepisosteus_oculatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSLAFG00000011082 | RRAGA | 100 | 97.764 | Loxodonta_africana |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSMFAG00000041833 | RRAGB | 100 | 99.422 | Macaca_fascicularis |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMFAG00000030433 | RRAGA | 100 | 98.083 | Macaca_fascicularis |
| ENSG00000083750 | RRAGB | 98 | 99.476 | ENSMMUG00000017635 | RRAGB | 99 | 99.672 | Macaca_mulatta |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMMUG00000018395 | RRAGA | 99 | 91.040 | Macaca_mulatta |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMNEG00000019531 | RRAGA | 100 | 98.083 | Macaca_nemestrina |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMLEG00000025200 | RRAGA | 100 | 98.083 | Mandrillus_leucophaeus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSMLEG00000004971 | RRAGB | 100 | 99.422 | Mandrillus_leucophaeus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSMAMG00000012091 | rraga | 99 | 95.208 | Mastacembelus_armatus |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSMZEG00005015085 | rraga | 99 | 95.847 | Maylandia_zebra |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMGAG00000005575 | RRAGB | 100 | 97.764 | Meleagris_gallopavo |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMAUG00000003702 | Rraga | 100 | 98.083 | Mesocricetus_auratus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMICG00000016534 | RRAGA | 100 | 98.083 | Microcebus_murinus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSMICG00000035059 | RRAGB | 100 | 98.844 | Microcebus_murinus |
| ENSG00000083750 | RRAGB | 98 | 99.474 | ENSMOCG00000010568 | Rragb | 100 | 97.764 | Microtus_ochrogaster |
| ENSG00000083750 | RRAGB | 99 | 98.964 | ENSMODG00000011337 | RRAGB | 100 | 98.371 | Monodelphis_domestica |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSMALG00000020662 | rraga | 99 | 95.527 | Monopterus_albus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | MGP_CAROLIEiJ_G0026131 | Rraga | 100 | 98.083 | Mus_caroli |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMUSG00000070934 | Rraga | 100 | 98.083 | Mus_musculus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | MGP_PahariEiJ_G0028469 | Rraga | 100 | 98.083 | Mus_pahari |
| ENSG00000083750 | RRAGB | 100 | 98.969 | MGP_SPRETEiJ_G0027096 | Rraga | 100 | 98.083 | Mus_spretus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMPUG00000019121 | RRAGA | 100 | 98.083 | Mustela_putorius_furo |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSMLUG00000010870 | RRAGA | 100 | 98.083 | Myotis_lucifugus |
| ENSG00000083750 | RRAGB | 99 | 99.482 | ENSNGAG00000015964 | Rragb | 99 | 99.674 | Nannospalax_galili |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSNGAG00000007602 | Rraga | 100 | 98.083 | Nannospalax_galili |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSNBRG00000016166 | rraga | 99 | 95.847 | Neolamprologus_brichardi |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSNLEG00000016350 | RRAGB | 100 | 99.711 | Nomascus_leucogenys |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSNLEG00000018985 | RRAGA | 100 | 98.083 | Nomascus_leucogenys |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSOPRG00000005886 | RRAGA | 100 | 98.083 | Ochotona_princeps |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSODEG00000020270 | RRAGA | 100 | 97.764 | Octodon_degus |
| ENSG00000083750 | RRAGB | 98 | 98.429 | ENSODEG00000002951 | RRAGB | 99 | 96.215 | Octodon_degus |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSONIG00000004447 | rraga | 99 | 95.847 | Oreochromis_niloticus |
| ENSG00000083750 | RRAGB | 95 | 98.370 | ENSOANG00000031844 | RRAGB | 89 | 97.354 | Ornithorhynchus_anatinus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSOCUG00000003506 | RRAGA | 100 | 98.083 | Oryctolagus_cuniculus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSORLG00000009278 | rraga | 99 | 95.847 | Oryzias_latipes |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSORLG00020001597 | rraga | 99 | 95.847 | Oryzias_latipes_hni |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSORLG00015005697 | rraga | 99 | 95.847 | Oryzias_latipes_hsok |
| ENSG00000083750 | RRAGB | 100 | 96.392 | ENSOMEG00000005139 | rraga | 99 | 96.013 | Oryzias_melastigma |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSOARG00000008644 | RRAGA | 100 | 98.089 | Ovis_aries |
| ENSG00000083750 | RRAGB | 100 | 99.485 | ENSOARG00000005619 | RRAGB | 100 | 94.220 | Ovis_aries |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPPAG00000034614 | RRAGB | 100 | 100.000 | Pan_paniscus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPPAG00000012463 | RRAGA | 100 | 98.083 | Pan_paniscus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPPRG00000021943 | RRAGB | 100 | 98.844 | Panthera_pardus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPPRG00000021236 | RRAGA | 100 | 98.083 | Panthera_pardus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPTIG00000005300 | RRAGB | 97 | 98.802 | Panthera_tigris_altaica |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPTRG00000020798 | RRAGA | 99 | 92.420 | Pan_troglodytes |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPTRG00000021955 | RRAGB | 100 | 100.000 | Pan_troglodytes |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPANG00000003594 | RRAGB | 100 | 99.422 | Papio_anubis |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPANG00000000085 | RRAGA | 100 | 98.083 | Papio_anubis |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSPKIG00000021851 | rraga | 99 | 95.847 | Paramormyrops_kingsleyae |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPSIG00000012621 | RRAGB | 100 | 97.764 | Pelodiscus_sinensis |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSPMGG00000009530 | rraga | 99 | 95.208 | Periophthalmus_magnuspinnatus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPCIG00000010545 | RRAGB | 95 | 96.262 | Phascolarctos_cinereus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSPFOG00000019453 | rraga | 98 | 95.208 | Poecilia_formosa |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSPLAG00000006541 | rraga | 99 | 95.208 | Poecilia_latipinna |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSPMEG00000008057 | rraga | 99 | 95.208 | Poecilia_mexicana |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSPREG00000021154 | rraga | 99 | 95.208 | Poecilia_reticulata |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPPYG00000019196 | RRAGA | 100 | 98.083 | Pongo_abelii |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPPYG00000020398 | RRAGB | 100 | 99.711 | Pongo_abelii |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPCAG00000003962 | RRAGA | 100 | 98.083 | Procavia_capensis |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSPCOG00000009451 | RRAGB | 100 | 99.133 | Propithecus_coquereli |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSPCOG00000013740 | RRAGA | 100 | 98.083 | Propithecus_coquereli |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSPNYG00000021130 | rraga | 99 | 95.847 | Pundamilia_nyererei |
| ENSG00000083750 | RRAGB | 100 | 88.660 | ENSPNAG00000011028 | rraga | 99 | 91.054 | Pygocentrus_nattereri |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSRNOG00000007051 | Rraga | 100 | 98.083 | Rattus_norvegicus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSRBIG00000003572 | RRAGA | 100 | 98.083 | Rhinopithecus_bieti |
| ENSG00000083750 | RRAGB | 87 | 98.817 | ENSRBIG00000031864 | RRAGB | 94 | 86.624 | Rhinopithecus_bieti |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSRROG00000002426 | RRAGA | 100 | 98.083 | Rhinopithecus_roxellana |
| ENSG00000083750 | RRAGB | 99 | 61.538 | YML121W | - | 98 | 51.948 | Saccharomyces_cerevisiae |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSSBOG00000012810 | RRAGB | 100 | 99.422 | Saimiri_boliviensis_boliviensis |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSSBOG00000008170 | - | 100 | 98.083 | Saimiri_boliviensis_boliviensis |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSSHAG00000001266 | - | 94 | 96.262 | Sarcophilus_harrisii |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSSFOG00015011428 | rraga | 99 | 95.527 | Scleropages_formosus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSSMAG00000017797 | rraga | 99 | 95.527 | Scophthalmus_maximus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSSDUG00000014535 | rraga | 99 | 95.527 | Seriola_dumerili |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSSLDG00000014834 | rraga | 99 | 95.527 | Seriola_lalandi_dorsalis |
| ENSG00000083750 | RRAGB | 97 | 97.884 | ENSSPUG00000019253 | RRAGB | 100 | 96.382 | Sphenodon_punctatus |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSSPAG00000009979 | rraga | 99 | 95.527 | Stegastes_partitus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSSSCG00000005174 | RRAGA | 100 | 98.083 | Sus_scrofa |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSSSCG00000037228 | RRAGB | 100 | 99.695 | Sus_scrofa |
| ENSG00000083750 | RRAGB | 100 | 98.454 | ENSTGUG00000005543 | RRAGB | 100 | 97.452 | Taeniopygia_guttata |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSTRUG00000013796 | rraga | 99 | 94.888 | Takifugu_rubripes |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSTNIG00000011814 | rraga | 99 | 95.208 | Tetraodon_nigroviridis |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSUAMG00000001776 | RRAGA | 98 | 98.680 | Ursus_americanus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSUAMG00000004721 | RRAGB | 100 | 98.844 | Ursus_americanus |
| ENSG00000083750 | RRAGB | 100 | 100.000 | ENSUMAG00000024099 | RRAGB | 100 | 98.844 | Ursus_maritimus |
| ENSG00000083750 | RRAGB | 100 | 98.969 | ENSVVUG00000000604 | RRAGA | 98 | 92.669 | Vulpes_vulpes |
| ENSG00000083750 | RRAGB | 100 | 99.485 | ENSVVUG00000015486 | RRAGB | 84 | 98.089 | Vulpes_vulpes |
| ENSG00000083750 | RRAGB | 100 | 97.423 | ENSXETG00000000887 | rraga | 100 | 96.486 | Xenopus_tropicalis |
| ENSG00000083750 | RRAGB | 100 | 96.907 | ENSXMAG00000012105 | rraga | 99 | 95.208 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0003924 | GTPase activity | 23723238. | IDA | Function |
| GO:0005515 | protein binding | 11073942.20381137.22424946.22575674.23394946.25259925.25561175.25567906. | IPI | Function |
| GO:0005525 | GTP binding | 7499430. | IDA | Function |
| GO:0005525 | GTP binding | 22424946.23723238. | IMP | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 9394008. | IDA | Component |
| GO:0005764 | lysosome | 21873635. | IBA | Component |
| GO:0005764 | lysosome | 20381137. | IDA | Component |
| GO:0005765 | lysosomal membrane | 17897319. | HDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007050 | cell cycle arrest | - | TAS | Process |
| GO:0009267 | cellular response to starvation | 21873635. | IBA | Process |
| GO:0010506 | regulation of autophagy | 21873635. | IBA | Process |
| GO:0016241 | regulation of macroautophagy | - | TAS | Process |
| GO:0032006 | regulation of TOR signaling | 23723238. | IGI | Process |
| GO:0032008 | positive regulation of TOR signaling | 21873635. | IBA | Process |
| GO:0032008 | positive regulation of TOR signaling | 22575674. | IMP | Process |
| GO:0032561 | guanyl ribonucleotide binding | 22575674. | IDA | Function |
| GO:0034198 | cellular response to amino acid starvation | 23723238. | IGI | Process |
| GO:0034448 | EGO complex | 21873635. | IBA | Component |
| GO:0034613 | cellular protein localization | 20381137. | IMP | Process |
| GO:0046982 | protein heterodimerization activity | 22424946. | IMP | Function |
| GO:0051020 | GTPase binding | 22424946. | IPI | Function |
| GO:0071230 | cellular response to amino acid stimulus | 21873635. | IBA | Process |
| GO:0071230 | cellular response to amino acid stimulus | 20381137. | IMP | Process |
| GO:1904263 | positive regulation of TORC1 signaling | 22424946. | IMP | Process |
| GO:1990131 | Gtr1-Gtr2 GTPase complex | 21873635. | IBA | Component |
| GO:1990253 | cellular response to leucine starvation | 22424946. | IMP | Process |