
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000372759 | ZMPSTE24-201 | 3108 | - | ENSP00000361845 | 475 (aa) | - | O75844 |
| ENST00000474142 | ZMPSTE24-203 | 530 | - | - | - (aa) | - | - |
| ENST00000472583 | ZMPSTE24-202 | 537 | - | - | - (aa) | - | - |
| ENST00000479131 | ZMPSTE24-204 | 561 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00900 | Terpenoid backbone biosynthesis | KEGG |
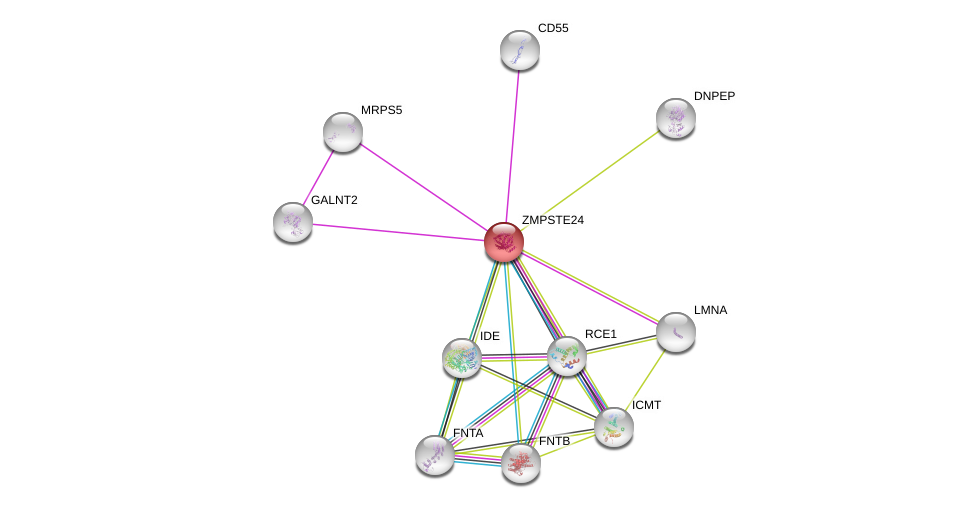
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001889 | liver development | - | IEA | Process |
| GO:0001942 | hair follicle development | - | IEA | Process |
| GO:0003007 | heart morphogenesis | - | IEA | Process |
| GO:0003229 | ventricular cardiac muscle tissue development | - | IEA | Process |
| GO:0003231 | cardiac ventricle development | - | IEA | Process |
| GO:0003417 | growth plate cartilage development | - | IEA | Process |
| GO:0003690 | double-stranded DNA binding | - | IEA | Function |
| GO:0004222 | metalloendopeptidase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 23539603.28169297. | IPI | Function |
| GO:0005637 | nuclear inner membrane | - | IEA | Component |
| GO:0006281 | DNA repair | - | IEA | Process |
| GO:0006508 | proteolysis | 10373325. | TAS | Process |
| GO:0006925 | inflammatory cell apoptotic process | - | IEA | Process |
| GO:0006998 | nuclear envelope organization | - | IEA | Process |
| GO:0007628 | adult walking behavior | - | IEA | Process |
| GO:0008235 | metalloexopeptidase activity | 10373325. | TAS | Function |
| GO:0008340 | determination of adult lifespan | - | IEA | Process |
| GO:0008360 | regulation of cell shape | - | IEA | Process |
| GO:0010506 | regulation of autophagy | - | IEA | Process |
| GO:0010906 | regulation of glucose metabolic process | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0019216 | regulation of lipid metabolic process | - | IEA | Process |
| GO:0030176 | integral component of endoplasmic reticulum membrane | 21873635. | IBA | Component |
| GO:0030282 | bone mineralization | - | IEA | Process |
| GO:0030327 | prenylated protein catabolic process | - | IEA | Process |
| GO:0030500 | regulation of bone mineralization | - | IEA | Process |
| GO:0032006 | regulation of TOR signaling | - | IEA | Process |
| GO:0032350 | regulation of hormone metabolic process | - | IEA | Process |
| GO:0032991 | protein-containing complex | 28246125. | IDA | Component |
| GO:0035264 | multicellular organism growth | - | IEA | Process |
| GO:0040014 | regulation of multicellular organism growth | - | IEA | Process |
| GO:0043007 | maintenance of rDNA | - | IEA | Process |
| GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | - | IEA | Process |
| GO:0043979 | histone H2B-K5 acetylation | - | IEA | Process |
| GO:0044029 | hypomethylation of CpG island | - | IEA | Process |
| GO:0044255 | cellular lipid metabolic process | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0048145 | regulation of fibroblast proliferation | - | IEA | Process |
| GO:0048538 | thymus development | - | IEA | Process |
| GO:0048739 | cardiac muscle fiber development | - | IEA | Process |
| GO:0050688 | regulation of defense response to virus | 28246125. | IDA | Process |
| GO:0050905 | neuromuscular process | - | IEA | Process |
| GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | - | IEA | Process |
| GO:0060993 | kidney morphogenesis | - | IEA | Process |
| GO:0061337 | cardiac conduction | - | IEA | Process |
| GO:0061762 | CAMKK-AMPK signaling cascade | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0070302 | regulation of stress-activated protein kinase signaling cascade | - | IEA | Process |
| GO:0071480 | cellular response to gamma radiation | - | IEA | Process |
| GO:0071586 | CAAX-box protein processing | 21873635. | IBA | Process |
| GO:0072423 | response to DNA damage checkpoint signaling | - | IEA | Process |
| GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | - | IEA | Process |
| GO:1903463 | regulation of mitotic cell cycle DNA replication | - | IEA | Process |
| GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA | - | IEA | Process |
| GO:1990036 | calcium ion import into sarcoplasmic reticulum | - | IEA | Process |
| GO:1990164 | histone H2A phosphorylation | - | IEA | Process |
| GO:2000618 | regulation of histone H4-K16 acetylation | - | IEA | Process |
| GO:2000730 | regulation of termination of RNA polymerase I transcription | - | IEA | Process |
| GO:2000772 | regulation of cellular senescence | - | IEA | Process |