
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000536078 | ADD1-221 | 423 | - | - | - (aa) | - | - |
| ENST00000264758 | ADD1-201 | 4045 | XM_005247934 | ENSP00000264758 | 768 (aa) | XP_005247991 | P35611 |
| ENST00000508277 | ADD1-212 | 491 | - | ENSP00000426700 | 142 (aa) | - | D6RF25 |
| ENST00000539108 | ADD1-224 | 244 | - | - | - (aa) | - | - |
| ENST00000540541 | ADD1-226 | 546 | - | - | - (aa) | - | - |
| ENST00000506157 | ADD1-211 | 5179 | - | - | - (aa) | - | - |
| ENST00000514940 | ADD1-219 | 1332 | - | ENSP00000424143 | 444 (aa) | - | H0Y9H2 |
| ENST00000511797 | ADD1-216 | 671 | - | ENSP00000421918 | 137 (aa) | - | D6RAH3 |
| ENST00000513762 | ADD1-218 | 3581 | - | - | - (aa) | - | - |
| ENST00000539149 | ADD1-225 | 460 | - | - | - (aa) | - | - |
| ENST00000541843 | ADD1-228 | 406 | - | ENSP00000441388 | 114 (aa) | - | H0YG19 |
| ENST00000510101 | ADD1-215 | 704 | - | ENSP00000425703 | 69 (aa) | - | D6RJE2 |
| ENST00000538860 | ADD1-223 | 507 | - | - | - (aa) | - | - |
| ENST00000355842 | ADD1-202 | 4743 | - | ENSP00000348100 | 662 (aa) | - | P35611 |
| ENST00000374281 | ADD1-203 | 445 | - | - | - (aa) | - | - |
| ENST00000503169 | ADD1-209 | 2394 | - | - | - (aa) | - | - |
| ENST00000503062 | ADD1-208 | 488 | - | - | - (aa) | - | - |
| ENST00000508684 | ADD1-213 | 3059 | - | - | - (aa) | - | - |
| ENST00000503455 | ADD1-210 | 3206 | - | ENSP00000423024 | 663 (aa) | - | E7ENY0 |
| ENST00000509039 | ADD1-214 | 1569 | - | - | - (aa) | - | - |
| ENST00000398129 | ADD1-206 | 3787 | XM_017007705 | ENSP00000381197 | 737 (aa) | XP_016863194 | P35611 |
| ENST00000513328 | ADD1-217 | 3107 | XM_017007709 | ENSP00000421907 | 632 (aa) | XP_016863198 | E7EV99 |
| ENST00000398123 | ADD1-204 | 2361 | XM_024453887 | ENSP00000381191 | 662 (aa) | XP_024309655 | P35611 |
| ENST00000446856 | ADD1-207 | 3745 | - | ENSP00000399828 | 400 (aa) | - | A0A0A0MSR2 |
| ENST00000398125 | ADD1-205 | 4079 | - | ENSP00000381193 | 662 (aa) | - | P35611 |
| ENST00000541051 | ADD1-227 | 975 | - | - | - (aa) | - | - |
| ENST00000536424 | ADD1-222 | 594 | - | ENSP00000438069 | 105 (aa) | - | H0YFD8 |
| ENST00000534870 | ADD1-220 | 795 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000087274 | Body Weights and Measures | 7.96492241699802E-6 | 17903300 |
| ENSG00000087274 | Blood Cells | 2.17464396275359E-6 | 17903294 |
| ENSG00000087274 | Alcohol Drinking | 2.3307513343843E-12 | - |
| ENSG00000087274 | Echocardiography | 1.08236443523306E-6 | 17903301 |
| ENSG00000087274 | Stroke | 8.8834442E-004 | - |
| ENSG00000087274 | Child Development Disorders, Pervasive | 2.8046000E-005 | - |
| ENSG00000087274 | Child Development Disorders, Pervasive | 4.4736000E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000087274 | rs6824567 | 4 | 2881445 | T | Educational attainment (MTAG) | 30038396 | [0.0094-0.0152] unit increase | 0.0123 | EFO_0004784 |
| ENSG00000087274 | rs114643193 | 4 | 2868202 | ? | Huntington's disease progression | 28642124 | Orphanet_399|EFO_0008336 | ||
| ENSG00000087274 | rs6824567 | 4 | 2881445 | T | Educational attainment (years of education) | 30038396 | [0.0099-0.0161] unit increase | 0.013 | EFO_0004784 |
| ENSG00000087274 | rs624833 | 4 | 2879529 | G | Age of smoking initiation (MTAG) | 30643251 | [0.0083-0.0145] unit increase | 0.0114003 | EFO_0005670 |
| ENSG00000087274 | rs624833 | 4 | 2879529 | G | Age of smoking initiation | 30643251 | [0.011-0.021] unit increase | 0.016 | EFO_0005670 |
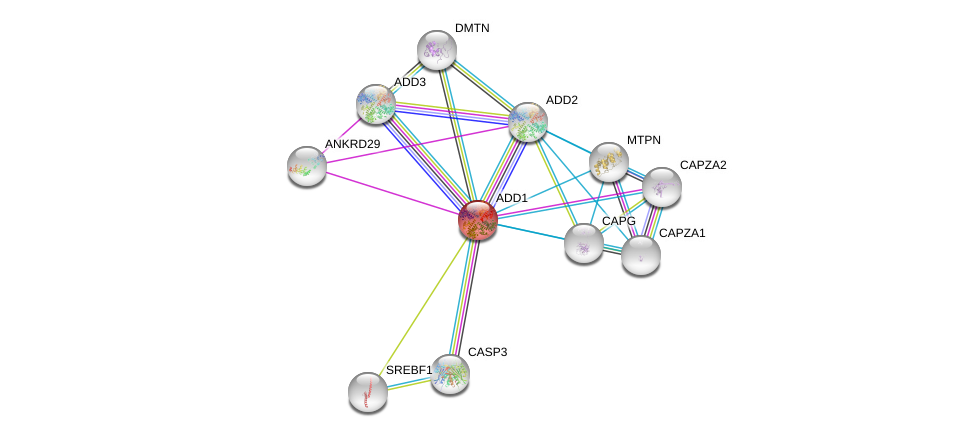
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000087274 | ADD1 | 100 | 98.462 | ENSMUSG00000029106 | Add1 | 100 | 97.872 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000902 | cell morphogenesis | - | IEA | Process |
| GO:0001701 | in utero embryonic development | - | IEA | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0003779 | actin binding | 1840603. | TAS | Function |
| GO:0005198 | structural molecule activity | 21873635. | IBA | Function |
| GO:0005516 | calmodulin binding | - | IEA | Function |
| GO:0005634 | nucleus | 16289097. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | 25978380. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005856 | cytoskeleton | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0005886 | plasma membrane | 25978380. | IDA | Component |
| GO:0005913 | cell-cell adherens junction | 21873635. | IBA | Component |
| GO:0005913 | cell-cell adherens junction | 25978380. | IDA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0006884 | cell volume homeostasis | - | IEA | Process |
| GO:0008134 | transcription factor binding | 16289097. | IPI | Function |
| GO:0008290 | F-actin capping protein complex | 8626479. | IDA | Component |
| GO:0014069 | postsynaptic density | 21873635. | IBA | Component |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0020027 | hemoglobin metabolic process | - | IEA | Process |
| GO:0030036 | actin cytoskeleton organization | 3693401. | IC | Process |
| GO:0030218 | erythrocyte differentiation | - | IEA | Process |
| GO:0030507 | spectrin binding | 21873635. | IBA | Function |
| GO:0030507 | spectrin binding | 7642559. | IDA | Function |
| GO:0032092 | positive regulation of protein binding | 3693401.7642559. | IDA | Process |
| GO:0035264 | multicellular organism growth | - | IEA | Process |
| GO:0036498 | IRE1-mediated unfolded protein response | - | TAS | Process |
| GO:0042803 | protein homodimerization activity | 7642559. | IPI | Function |
| GO:0044853 | plasma membrane raft | 21873635. | IBA | Component |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0046982 | protein heterodimerization activity | 7642559. | IPI | Function |
| GO:0048873 | homeostasis of number of cells within a tissue | - | IEA | Process |
| GO:0051015 | actin filament binding | 21873635. | IBA | Function |
| GO:0051015 | actin filament binding | 7642559. | IDA | Function |
| GO:0051016 | barbed-end actin filament capping | 21873635. | IBA | Process |
| GO:0051016 | barbed-end actin filament capping | 8626479. | IDA | Process |
| GO:0051017 | actin filament bundle assembly | 21873635. | IBA | Process |
| GO:0051017 | actin filament bundle assembly | 3693401. | IDA | Process |
| GO:0055085 | transmembrane transport | - | TAS | Process |
| GO:0071277 | cellular response to calcium ion | 25978380. | IDA | Process |
| GO:1903142 | positive regulation of establishment of endothelial barrier | 21873635. | IBA | Process |
| GO:1903142 | positive regulation of establishment of endothelial barrier | 25978380. | IDA | Process |
| GO:1903393 | positive regulation of adherens junction organization | 21873635. | IBA | Process |
| GO:1903393 | positive regulation of adherens junction organization | 25978380. | IDA | Process |