
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000408947 | 2OG-FeII_Oxy | PF03171.20 | 8.7e-09 | 1 | 1 |
| ENSP00000316881 | 2OG-FeII_Oxy | PF03171.20 | 1.5e-08 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000444866 | P3H2-204 | 544 | - | ENSP00000391374 | 137 (aa) | - | C9J313 |
| ENST00000482780 | P3H2-209 | 893 | - | - | - (aa) | - | - |
| ENST00000463171 | P3H2-205 | 754 | - | - | - (aa) | - | - |
| ENST00000475095 | P3H2-208 | 552 | - | - | - (aa) | - | - |
| ENST00000427335 | P3H2-203 | 2963 | XM_011512955 | ENSP00000408947 | 527 (aa) | XP_011511257 | Q8IVL5 |
| ENST00000467131 | P3H2-206 | 554 | - | - | - (aa) | - | - |
| ENST00000319332 | P3H2-201 | 3509 | - | ENSP00000316881 | 708 (aa) | - | Q8IVL5 |
| ENST00000470925 | P3H2-207 | 583 | - | - | - (aa) | - | - |
| ENST00000490940 | P3H2-210 | 472 | - | - | - (aa) | - | - |
| ENST00000426003 | P3H2-202 | 609 | - | ENSP00000394326 | 134 (aa) | - | C9JSL4 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000090530 | Blood Pressure | 8.92867062781078E-5 | 17903302 |
| ENSG00000090530 | Blood Pressure | 7.6855409045791E-5 | 17903302 |
| ENSG00000090530 | Blood Pressure | 7.78253244468834E-5 | 17903302 |
| ENSG00000090530 | Transferrin | 7.557e-005 | - |
| ENSG00000090530 | Calcium | 9.6780000E-005 | - |
| ENSG00000090530 | Blood Pressure | 2.9863061E-005 | - |
| ENSG00000090530 | Platelet Function Tests | 2.6693100E-005 | - |
| ENSG00000090530 | Platelet Function Tests | 1.6268000E-005 | - |
| ENSG00000090530 | Platelet Function Tests | 1.7823000E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000090530 | rs13319406 | 3 | 190118647 | G | Itch intensity from mosquito bite adjusted by bite size | 28199695 | [0.019-0.046] unit increase | 0.0324478 | EFO_0008377|EFO_0008378 |
| ENSG00000090530 | rs2030571 | 3 | 190104301 | T | Facial morphology (factor 16) | 28441456 | [0.18-0.43] unit increase | 0.3081 | EFO_0007841 |
| ENSG00000090530 | rs3106806 | 3 | 190103894 | C | Nicotine dependence and major depression (severity of comorbidity) | 30287806 | [0.74-1.81] unit decrease | 1.2707 | EFO_0009262|EFO_0007006 |
| ENSG00000090530 | rs9882792 | 3 | 190056667 | T | Breast cancer | 29059683 | [0.022-0.053] unit decrease | 0.0377 | EFO_0000305 |
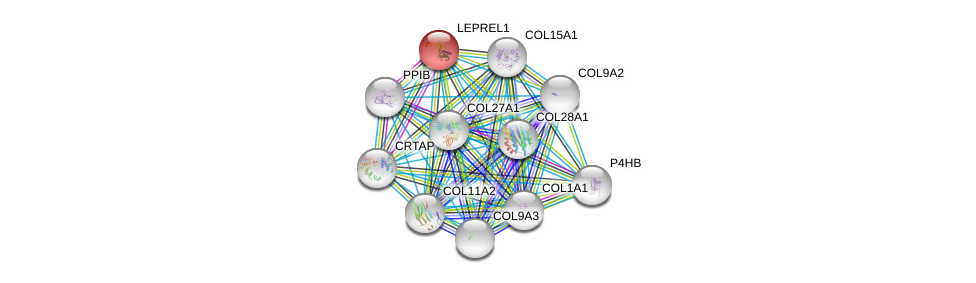
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000090530 | P3H2 | 99 | 62.667 | ENSAPOG00000021463 | p3h2 | 100 | 62.667 | Acanthochromis_polyacanthus |
| ENSG00000090530 | P3H2 | 100 | 90.892 | ENSAMEG00000000726 | P3H2 | 100 | 89.661 | Ailuropoda_melanoleuca |
| ENSG00000090530 | P3H2 | 100 | 62.429 | ENSACIG00000012482 | p3h2 | 96 | 59.701 | Amphilophus_citrinellus |
| ENSG00000090530 | P3H2 | 83 | 65.217 | ENSAOCG00000006595 | p3h2 | 96 | 65.217 | Amphiprion_ocellaris |
| ENSG00000090530 | P3H2 | 100 | 61.860 | ENSAPEG00000006169 | p3h2 | 96 | 59.281 | Amphiprion_percula |
| ENSG00000090530 | P3H2 | 100 | 62.239 | ENSATEG00000009514 | p3h2 | 96 | 59.132 | Anabas_testudineus |
| ENSG00000090530 | P3H2 | 100 | 96.774 | ENSANAG00000021430 | P3H2 | 100 | 94.640 | Aotus_nancymaae |
| ENSG00000090530 | P3H2 | 100 | 61.480 | ENSACLG00000020074 | p3h2 | 96 | 59.104 | Astatotilapia_calliptera |
| ENSG00000090530 | P3H2 | 100 | 62.619 | ENSAMXG00000017679 | p3h2 | 96 | 58.832 | Astyanax_mexicanus |
| ENSG00000090530 | P3H2 | 100 | 95.636 | ENSBTAG00000021025 | P3H2 | 100 | 92.655 | Bos_taurus |
| ENSG00000090530 | P3H2 | 100 | 97.154 | ENSCJAG00000014239 | P3H2 | 100 | 94.789 | Callithrix_jacchus |
| ENSG00000090530 | P3H2 | 100 | 94.877 | ENSCAFG00000013985 | P3H2 | 100 | 91.139 | Canis_familiaris |
| ENSG00000090530 | P3H2 | 100 | 94.877 | ENSCAFG00020020127 | P3H2 | 100 | 91.139 | Canis_lupus_dingo |
| ENSG00000090530 | P3H2 | 100 | 93.928 | ENSCHIG00000011298 | P3H2 | 100 | 91.384 | Capra_hircus |
| ENSG00000090530 | P3H2 | 100 | 96.774 | ENSTSYG00000008652 | P3H2 | 100 | 96.774 | Carlito_syrichta |
| ENSG00000090530 | P3H2 | 100 | 87.591 | ENSCAPG00000012826 | P3H2 | 100 | 79.091 | Cavia_aperea |
| ENSG00000090530 | P3H2 | 100 | 90.892 | ENSCPOG00000004329 | P3H2 | 95 | 90.182 | Cavia_porcellus |
| ENSG00000090530 | P3H2 | 100 | 97.154 | ENSCCAG00000034046 | P3H2 | 100 | 94.225 | Cebus_capucinus |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSCATG00000004107 | P3H2 | 100 | 98.023 | Cercocebus_atys |
| ENSG00000090530 | P3H2 | 100 | 92.410 | ENSCLAG00000012801 | P3H2 | 100 | 90.500 | Chinchilla_lanigera |
| ENSG00000090530 | P3H2 | 100 | 98.672 | ENSCSAG00000009034 | P3H2 | 100 | 98.023 | Chlorocebus_sabaeus |
| ENSG00000090530 | P3H2 | 82 | 97.368 | ENSCHOG00000013468 | P3H2 | 82 | 97.368 | Choloepus_hoffmanni |
| ENSG00000090530 | P3H2 | 100 | 78.748 | ENSCPBG00000012789 | P3H2 | 97 | 78.832 | Chrysemys_picta_bellii |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSCANG00000017978 | P3H2 | 99 | 98.851 | Colobus_angolensis_palliatus |
| ENSG00000090530 | P3H2 | 100 | 92.979 | ENSCGRG00001021764 | P3h2 | 97 | 90.393 | Cricetulus_griseus_chok1gshd |
| ENSG00000090530 | P3H2 | 100 | 92.789 | ENSCGRG00000009640 | P3h2 | 98 | 92.219 | Cricetulus_griseus_crigri |
| ENSG00000090530 | P3H2 | 100 | 62.239 | ENSCSEG00000013736 | p3h2 | 95 | 59.880 | Cynoglossus_semilaevis |
| ENSG00000090530 | P3H2 | 100 | 61.670 | ENSCVAG00000020470 | p3h2 | 96 | 58.383 | Cyprinodon_variegatus |
| ENSG00000090530 | P3H2 | 100 | 62.239 | ENSDARG00000103026 | p3h2 | 100 | 62.286 | Danio_rerio |
| ENSG00000090530 | P3H2 | 100 | 94.118 | ENSDNOG00000035205 | P3H2 | 100 | 90.819 | Dasypus_novemcinctus |
| ENSG00000090530 | P3H2 | 100 | 93.169 | ENSDORG00000001518 | P3h2 | 100 | 90.704 | Dipodomys_ordii |
| ENSG00000090530 | P3H2 | 100 | 95.446 | ENSEASG00005000187 | P3H2 | 100 | 91.949 | Equus_asinus_asinus |
| ENSG00000090530 | P3H2 | 100 | 96.015 | ENSECAG00000008971 | P3H2 | 100 | 92.514 | Equus_caballus |
| ENSG00000090530 | P3H2 | 100 | 96.015 | ENSFCAG00000000791 | P3H2 | 100 | 92.948 | Felis_catus |
| ENSG00000090530 | P3H2 | 100 | 71.904 | ENSFALG00000007377 | P3H2 | 100 | 70.256 | Ficedula_albicollis |
| ENSG00000090530 | P3H2 | 100 | 63.188 | ENSFHEG00000007209 | p3h2 | 95 | 60.030 | Fundulus_heteroclitus |
| ENSG00000090530 | P3H2 | 100 | 61.815 | ENSGMOG00000009134 | p3h2 | 100 | 61.815 | Gadus_morhua |
| ENSG00000090530 | P3H2 | 100 | 78.368 | ENSGALG00000040866 | P3H2 | 96 | 75.373 | Gallus_gallus |
| ENSG00000090530 | P3H2 | 100 | 62.808 | ENSGAFG00000013603 | p3h2 | 96 | 60.180 | Gambusia_affinis |
| ENSG00000090530 | P3H2 | 100 | 62.075 | ENSGACG00000010066 | p3h2 | 100 | 62.075 | Gasterosteus_aculeatus |
| ENSG00000090530 | P3H2 | 100 | 78.748 | ENSGAGG00000025337 | - | 86 | 74.707 | Gopherus_agassizii |
| ENSG00000090530 | P3H2 | 100 | 99.431 | ENSGGOG00000009749 | P3H2 | 100 | 98.596 | Gorilla_gorilla |
| ENSG00000090530 | P3H2 | 99 | 56.061 | ENSHBUG00000022967 | p3h2 | 100 | 54.753 | Haplochromis_burtoni |
| ENSG00000090530 | P3H2 | 100 | 90.702 | ENSHGLG00000019175 | P3H2 | 97 | 87.083 | Heterocephalus_glaber_female |
| ENSG00000090530 | P3H2 | 100 | 56.204 | ENSHCOG00000019274 | p3h2 | 97 | 53.892 | Hippocampus_comes |
| ENSG00000090530 | P3H2 | 99 | 60.381 | ENSIPUG00000016970 | p3h2 | 99 | 56.560 | Ictalurus_punctatus |
| ENSG00000090530 | P3H2 | 100 | 94.497 | ENSSTOG00000006501 | P3H2 | 100 | 91.243 | Ictidomys_tridecemlineatus |
| ENSG00000090530 | P3H2 | 100 | 91.651 | ENSJJAG00000013941 | P3h2 | 95 | 91.289 | Jaculus_jaculus |
| ENSG00000090530 | P3H2 | 100 | 62.998 | ENSKMAG00000022098 | p3h2 | 98 | 59.763 | Kryptolebias_marmoratus |
| ENSG00000090530 | P3H2 | 100 | 62.998 | ENSLBEG00000003942 | p3h2 | 98 | 59.232 | Labrus_bergylta |
| ENSG00000090530 | P3H2 | 100 | 69.754 | ENSLACG00000014642 | P3H2 | 96 | 65.522 | Latimeria_chalumnae |
| ENSG00000090530 | P3H2 | 100 | 65.844 | ENSLOCG00000005129 | p3h2 | 96 | 62.575 | Lepisosteus_oculatus |
| ENSG00000090530 | P3H2 | 100 | 93.738 | ENSLAFG00000007727 | P3H2 | 100 | 93.333 | Loxodonta_africana |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSMFAG00000036104 | P3H2 | 100 | 98.023 | Macaca_fascicularis |
| ENSG00000090530 | P3H2 | 100 | 98.672 | ENSMMUG00000000623 | P3H2 | 100 | 97.881 | Macaca_mulatta |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSMNEG00000027261 | P3H2 | 100 | 98.023 | Macaca_nemestrina |
| ENSG00000090530 | P3H2 | 99 | 61.524 | ENSMAMG00000006370 | p3h2 | 100 | 61.524 | Mastacembelus_armatus |
| ENSG00000090530 | P3H2 | 100 | 61.480 | ENSMZEG00005003110 | p3h2 | 96 | 59.104 | Maylandia_zebra |
| ENSG00000090530 | P3H2 | 100 | 77.609 | ENSMGAG00000007491 | P3H2 | 100 | 77.413 | Meleagris_gallopavo |
| ENSG00000090530 | P3H2 | 100 | 92.600 | ENSMAUG00000004803 | P3h2 | 97 | 89.956 | Mesocricetus_auratus |
| ENSG00000090530 | P3H2 | 100 | 96.964 | ENSMICG00000049208 | P3H2 | 100 | 93.644 | Microcebus_murinus |
| ENSG00000090530 | P3H2 | 100 | 93.359 | ENSMOCG00000008061 | P3h2 | 97 | 90.830 | Microtus_ochrogaster |
| ENSG00000090530 | P3H2 | 100 | 61.290 | ENSMMOG00000019576 | p3h2 | 96 | 58.832 | Mola_mola |
| ENSG00000090530 | P3H2 | 97 | 68.750 | ENSMODG00000015073 | P3H2 | 95 | 63.529 | Monodelphis_domestica |
| ENSG00000090530 | P3H2 | 100 | 62.049 | ENSMALG00000017825 | p3h2 | 96 | 58.683 | Monopterus_albus |
| ENSG00000090530 | P3H2 | 100 | 92.220 | MGP_CAROLIEiJ_G0020626 | P3h2 | 97 | 90.102 | Mus_caroli |
| ENSG00000090530 | P3H2 | 100 | 92.220 | ENSMUSG00000038168 | P3h2 | 97 | 90.102 | Mus_musculus |
| ENSG00000090530 | P3H2 | 100 | 93.169 | MGP_PahariEiJ_G0016331 | P3h2 | 97 | 91.121 | Mus_pahari |
| ENSG00000090530 | P3H2 | 100 | 92.030 | MGP_SPRETEiJ_G0021522 | P3h2 | 97 | 89.942 | Mus_spretus |
| ENSG00000090530 | P3H2 | 99 | 95.812 | ENSMPUG00000003446 | P3H2 | 88 | 95.812 | Mustela_putorius_furo |
| ENSG00000090530 | P3H2 | 100 | 93.750 | ENSMLUG00000015613 | P3H2 | 100 | 93.455 | Myotis_lucifugus |
| ENSG00000090530 | P3H2 | 100 | 92.410 | ENSNGAG00000000626 | P3h2 | 98 | 85.358 | Nannospalax_galili |
| ENSG00000090530 | P3H2 | 100 | 59.583 | ENSNBRG00000016641 | p3h2 | 97 | 57.463 | Neolamprologus_brichardi |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSNLEG00000006734 | - | 100 | 98.861 | Nomascus_leucogenys |
| ENSG00000090530 | P3H2 | 100 | 93.169 | ENSOPRG00000014074 | P3H2 | 100 | 89.873 | Ochotona_princeps |
| ENSG00000090530 | P3H2 | 100 | 61.101 | ENSONIG00000001714 | p3h2 | 96 | 58.358 | Oreochromis_niloticus |
| ENSG00000090530 | P3H2 | 94 | 85.170 | ENSOANG00000013797 | P3H2 | 99 | 85.170 | Ornithorhynchus_anatinus |
| ENSG00000090530 | P3H2 | 100 | 94.687 | ENSOCUG00000011014 | P3H2 | 100 | 91.842 | Oryctolagus_cuniculus |
| ENSG00000090530 | P3H2 | 100 | 61.101 | ENSORLG00000005641 | p3h2 | 96 | 58.383 | Oryzias_latipes |
| ENSG00000090530 | P3H2 | 100 | 56.716 | ENSORLG00020021674 | p3h2 | 95 | 57.878 | Oryzias_latipes_hni |
| ENSG00000090530 | P3H2 | 100 | 61.290 | ENSORLG00015019012 | p3h2 | 96 | 58.533 | Oryzias_latipes_hsok |
| ENSG00000090530 | P3H2 | 100 | 61.670 | ENSOMEG00000013713 | p3h2 | 96 | 59.132 | Oryzias_melastigma |
| ENSG00000090530 | P3H2 | 100 | 93.585 | ENSOGAG00000003842 | P3H2 | 100 | 93.466 | Otolemur_garnettii |
| ENSG00000090530 | P3H2 | 100 | 93.785 | ENSOARG00000020475 | P3H2 | 100 | 91.292 | Ovis_aries |
| ENSG00000090530 | P3H2 | 100 | 99.431 | ENSPPAG00000040013 | P3H2 | 95 | 99.506 | Pan_paniscus |
| ENSG00000090530 | P3H2 | 100 | 95.825 | ENSPPRG00000014656 | P3H2 | 100 | 92.525 | Panthera_pardus |
| ENSG00000090530 | P3H2 | 100 | 95.825 | ENSPTIG00000016608 | P3H2 | 84 | 95.784 | Panthera_tigris_altaica |
| ENSG00000090530 | P3H2 | 100 | 99.435 | ENSPTRG00000015736 | P3H2 | 100 | 99.435 | Pan_troglodytes |
| ENSG00000090530 | P3H2 | 100 | 98.672 | ENSPANG00000005379 | P3H2 | 100 | 97.881 | Papio_anubis |
| ENSG00000090530 | P3H2 | 100 | 62.239 | ENSPMGG00000012834 | p3h2 | 97 | 59.431 | Periophthalmus_magnuspinnatus |
| ENSG00000090530 | P3H2 | 100 | 92.789 | ENSPEMG00000016547 | P3h2 | 97 | 90.379 | Peromyscus_maniculatus_bairdii |
| ENSG00000090530 | P3H2 | 100 | 63.378 | ENSPFOG00000014376 | p3h2 | 96 | 60.479 | Poecilia_formosa |
| ENSG00000090530 | P3H2 | 100 | 63.188 | ENSPLAG00000012556 | p3h2 | 96 | 60.329 | Poecilia_latipinna |
| ENSG00000090530 | P3H2 | 100 | 62.998 | ENSPMEG00000010685 | p3h2 | 95 | 60.180 | Poecilia_mexicana |
| ENSG00000090530 | P3H2 | 100 | 60.721 | ENSPREG00000015431 | p3h2 | 96 | 57.485 | Poecilia_reticulata |
| ENSG00000090530 | P3H2 | 100 | 94.118 | ENSPVAG00000009057 | P3H2 | 100 | 91.549 | Pteropus_vampyrus |
| ENSG00000090530 | P3H2 | 100 | 61.670 | ENSPNYG00000005890 | p3h2 | 96 | 59.254 | Pundamilia_nyererei |
| ENSG00000090530 | P3H2 | 100 | 62.619 | ENSPNAG00000029093 | p3h2 | 97 | 59.552 | Pygocentrus_nattereri |
| ENSG00000090530 | P3H2 | 100 | 92.979 | ENSRNOG00000055751 | P3h2 | 97 | 90.262 | Rattus_norvegicus |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSRBIG00000018185 | P3H2 | 100 | 98.164 | Rhinopithecus_bieti |
| ENSG00000090530 | P3H2 | 100 | 98.861 | ENSRROG00000031164 | P3H2 | 100 | 97.881 | Rhinopithecus_roxellana |
| ENSG00000090530 | P3H2 | 100 | 95.620 | ENSSBOG00000035056 | P3H2 | 100 | 93.453 | Saimiri_boliviensis_boliviensis |
| ENSG00000090530 | P3H2 | 96 | 66.535 | ENSSHAG00000006714 | - | 100 | 65.146 | Sarcophilus_harrisii |
| ENSG00000090530 | P3H2 | 100 | 63.757 | ENSSFOG00015006816 | p3h2 | 96 | 61.377 | Scleropages_formosus |
| ENSG00000090530 | P3H2 | 100 | 62.998 | ENSSMAG00000003284 | p3h2 | 95 | 60.329 | Scophthalmus_maximus |
| ENSG00000090530 | P3H2 | 100 | 63.567 | ENSSDUG00000002767 | p3h2 | 96 | 59.731 | Seriola_dumerili |
| ENSG00000090530 | P3H2 | 100 | 63.567 | ENSSLDG00000017365 | p3h2 | 96 | 59.731 | Seriola_lalandi_dorsalis |
| ENSG00000090530 | P3H2 | 100 | 70.896 | ENSSPUG00000010809 | P3H2 | 98 | 64.222 | Sphenodon_punctatus |
| ENSG00000090530 | P3H2 | 100 | 61.290 | ENSSPAG00000021635 | p3h2 | 96 | 58.533 | Stegastes_partitus |
| ENSG00000090530 | P3H2 | 100 | 95.446 | ENSSSCG00000011813 | P3H2 | 100 | 92.514 | Sus_scrofa |
| ENSG00000090530 | P3H2 | 100 | 76.850 | ENSTGUG00000009345 | P3H2 | 100 | 76.545 | Taeniopygia_guttata |
| ENSG00000090530 | P3H2 | 100 | 62.429 | ENSTRUG00000017655 | p3h2 | 96 | 59.581 | Takifugu_rubripes |
| ENSG00000090530 | P3H2 | 100 | 61.480 | ENSTNIG00000015540 | p3h2 | 100 | 61.566 | Tetraodon_nigroviridis |
| ENSG00000090530 | P3H2 | 100 | 87.591 | ENSTBEG00000004997 | P3H2 | 100 | 78.511 | Tupaia_belangeri |
| ENSG00000090530 | P3H2 | 100 | 92.220 | ENSTTRG00000001893 | P3H2 | 100 | 91.091 | Tursiops_truncatus |
| ENSG00000090530 | P3H2 | 100 | 91.971 | ENSUAMG00000006549 | P3H2 | 98 | 90.221 | Ursus_americanus |
| ENSG00000090530 | P3H2 | 100 | 95.825 | ENSUMAG00000003698 | P3H2 | 97 | 95.733 | Ursus_maritimus |
| ENSG00000090530 | P3H2 | 100 | 86.338 | ENSVPAG00000012351 | P3H2 | 100 | 86.903 | Vicugna_pacos |
| ENSG00000090530 | P3H2 | 100 | 94.877 | ENSVVUG00000013454 | P3H2 | 99 | 92.122 | Vulpes_vulpes |
| ENSG00000090530 | P3H2 | 100 | 69.565 | ENSXETG00000012845 | p3h2 | 100 | 69.550 | Xenopus_tropicalis |
| ENSG00000090530 | P3H2 | 100 | 61.101 | ENSXCOG00000002798 | p3h2 | 97 | 58.445 | Xiphophorus_couchianus |
| ENSG00000090530 | P3H2 | 100 | 62.239 | ENSXMAG00000018362 | p3h2 | 96 | 59.431 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | - | IEA | Function |
| GO:0005604 | basement membrane | - | ISS | Component |
| GO:0005783 | endoplasmic reticulum | 21873635. | IBA | Component |
| GO:0005783 | endoplasmic reticulum | 15063763. | IDA | Component |
| GO:0005788 | endoplasmic reticulum lumen | - | TAS | Component |
| GO:0005794 | Golgi apparatus | 15063763. | IDA | Component |
| GO:0008285 | negative regulation of cell proliferation | 19436308. | IDA | Process |
| GO:0016529 | sarcoplasmic reticulum | - | IEA | Component |
| GO:0019511 | peptidyl-proline hydroxylation | 18487197. | IDA | Process |
| GO:0019797 | procollagen-proline 3-dioxygenase activity | 21873635. | IBA | Function |
| GO:0019797 | procollagen-proline 3-dioxygenase activity | 18487197. | IDA | Function |
| GO:0031418 | L-ascorbic acid binding | - | IEA | Function |
| GO:0032963 | collagen metabolic process | 21873635. | IBA | Process |
| GO:0032963 | collagen metabolic process | 18487197. | IDA | Process |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PCPG | |||||||
| PCPG | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| KIRC | |||
| KIRP | |||
| LUAD | |||
| PAAD | |||
| SARC | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| MESO | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| ESCA | ||
| PAAD | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LGG |