
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000489184 | DLD-212 | 638 | - | - | - (aa) | - | - |
| ENST00000460577 | DLD-209 | 600 | - | - | - (aa) | - | - |
| ENST00000417551 | DLD-203 | 2041 | - | ENSP00000390667 | 509 (aa) | - | P09622 |
| ENST00000415325 | DLD-202 | 1802 | - | ENSP00000402593 | 58 (aa) | - | F8WDY5 |
| ENST00000453354 | DLD-208 | 537 | - | - | - (aa) | - | - |
| ENST00000478414 | DLD-210 | 549 | - | - | - (aa) | - | - |
| ENST00000485066 | DLD-211 | 483 | - | - | - (aa) | - | - |
| ENST00000494441 | DLD-213 | 707 | - | - | - (aa) | - | - |
| ENST00000451081 | DLD-207 | 930 | - | ENSP00000388077 | 115 (aa) | - | F2Z2E3 |
| ENST00000639772 | DLD-214 | 421 | - | ENSP00000492159 | 114 (aa) | - | A0A1W2PR83 |
| ENST00000437604 | DLD-204 | 1859 | - | ENSP00000387542 | 461 (aa) | - | P09622 |
| ENST00000205402 | DLD-201 | 3755 | - | ENSP00000205402 | 509 (aa) | - | P09622 |
| ENST00000450038 | DLD-206 | 682 | - | ENSP00000409590 | 114 (aa) | - | F8WDM5 |
| ENST00000440410 | DLD-205 | 1993 | - | ENSP00000417016 | 486 (aa) | - | E9PEX6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00010 | Glycolysis / Gluconeogenesis | KEGG |
| hsa00020 | Citrate cycle (TCA cycle) | KEGG |
| hsa00260 | Glycine, serine and threonine metabolism | KEGG |
| hsa00280 | Valine, leucine and isoleucine degradation | KEGG |
| hsa00310 | Lysine degradation | KEGG |
| hsa00380 | Tryptophan metabolism | KEGG |
| hsa00620 | Pyruvate metabolism | KEGG |
| hsa00630 | Glyoxylate and dicarboxylate metabolism | KEGG |
| hsa00640 | Propanoate metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
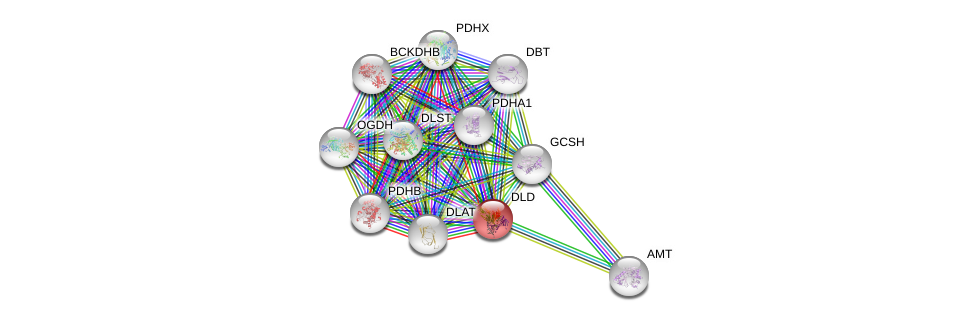
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000091140 | DLD | 100 | 94.892 | ENSMUSG00000020664 | Dld | 100 | 94.892 | Mus_musculus |
| ENSG00000091140 | DLD | 92 | 57.384 | YFL018C | LPD1 | 94 | 57.384 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004148 | dihydrolipoyl dehydrogenase activity | 16442803. | IDA | Function |
| GO:0005515 | protein binding | 16263718.16442803.29128334. | IPI | Function |
| GO:0005634 | nucleus | 29211711. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005739 | mitochondrion | 20833797. | HDA | Component |
| GO:0005739 | mitochondrion | 29211711. | IDA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0006090 | pyruvate metabolic process | - | TAS | Process |
| GO:0006099 | tricarboxylic acid cycle | - | TAS | Process |
| GO:0006103 | 2-oxoglutarate metabolic process | - | IEA | Process |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | - | IEA | Process |
| GO:0006508 | proteolysis | - | IEA | Process |
| GO:0006554 | lysine catabolic process | - | TAS | Process |
| GO:0007369 | gastrulation | - | IEA | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0009055 | electron transfer activity | - | IEA | Function |
| GO:0009083 | branched-chain amino acid catabolic process | - | TAS | Process |
| GO:0009106 | lipoate metabolic process | - | IEA | Process |
| GO:0031514 | motile cilium | - | IEA | Component |
| GO:0034604 | pyruvate dehydrogenase (NAD+) activity | 9242632. | IDA | Function |
| GO:0042391 | regulation of membrane potential | - | IEA | Process |
| GO:0043159 | acrosomal matrix | - | IEA | Component |
| GO:0043544 | lipoamide binding | - | IEA | Function |
| GO:0045252 | oxoglutarate dehydrogenase complex | 29211711. | IDA | Component |
| GO:0045254 | pyruvate dehydrogenase complex | 9242632. | IDA | Component |
| GO:0045454 | cell redox homeostasis | - | IEA | Process |
| GO:0048240 | sperm capacitation | - | IEA | Process |
| GO:0050660 | flavin adenine dinucleotide binding | - | IEA | Function |
| GO:0051068 | dihydrolipoamide metabolic process | - | IEA | Process |
| GO:0051287 | NAD binding | - | IEA | Function |
| GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate | 9242632. | IC | Process |
| GO:0106077 | histone succinylation | 29211711. | IDA | Process |