
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000452573 | RRM_1 | PF00076.22 | 2.9e-13 | 1 | 1 |
| ENSP00000451292 | RRM_1 | PF00076.22 | 3.2e-13 | 1 | 1 |
| ENSP00000450467 | RRM_1 | PF00076.22 | 3.6e-13 | 1 | 1 |
| ENSP00000452185 | RRM_1 | PF00076.22 | 7.1e-13 | 1 | 1 |
| ENSP00000452213 | RRM_1 | PF00076.22 | 8.1e-13 | 1 | 1 |
| ENSP00000450629 | RRM_1 | PF00076.22 | 9.7e-13 | 1 | 1 |
| ENSP00000452021 | RRM_1 | PF00076.22 | 1e-12 | 1 | 1 |
| ENSP00000451187 | RRM_1 | PF00076.22 | 1.1e-12 | 1 | 1 |
| ENSP00000452214 | RRM_1 | PF00076.22 | 1.1e-12 | 1 | 1 |
| ENSP00000404559 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000338095 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000450548 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000442816 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000450544 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000450725 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000451176 | RRM_1 | PF00076.22 | 1.3e-12 | 1 | 1 |
| ENSP00000404848 | RRM_1 | PF00076.22 | 1.4e-12 | 1 | 1 |
| ENSP00000451291 | RRM_1 | PF00076.22 | 1.4e-12 | 1 | 1 |
| ENSP00000452276 | RRM_1 | PF00076.22 | 1.4e-12 | 1 | 1 |
| ENSP00000451708 | RRM_1 | PF00076.22 | 1.5e-12 | 1 | 1 |
| ENSP00000450790 | RRM_1 | PF00076.22 | 1.7e-12 | 1 | 1 |
| ENSP00000451892 | RRM_1 | PF00076.22 | 2.3e-10 | 1 | 1 |
| ENSP00000450669 | RRM_1 | PF00076.22 | 3.7e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000553614 | HNRNPC-207 | 604 | - | - | - (aa) | - | - |
| ENST00000555127 | HNRNPC-215 | 929 | - | - | - (aa) | - | - |
| ENST00000556628 | HNRNPC-226 | 1156 | - | ENSP00000451652 | 226 (aa) | - | P07910 |
| ENST00000430246 | HNRNPC-203 | 4501 | - | ENSP00000442816 | 293 (aa) | - | P07910 |
| ENST00000557201 | HNRNPC-230 | 1371 | - | ENSP00000452276 | 306 (aa) | - | P07910 |
| ENST00000336053 | HNRNPC-201 | 3301 | - | ENSP00000338095 | 288 (aa) | - | B4DY08 |
| ENST00000557442 | HNRNPC-232 | 1308 | - | ENSP00000452599 | 147 (aa) | - | B4DSU6 |
| ENST00000555215 | HNRNPC-218 | 781 | - | ENSP00000452213 | 187 (aa) | - | G3V575 |
| ENST00000557768 | HNRNPC-233 | 599 | - | ENSP00000450669 | 54 (aa) | - | G3V2H6 |
| ENST00000554969 | HNRNPC-214 | 1924 | XM_017021253 | ENSP00000450725 | 293 (aa) | XP_016876742 | P07910 |
| ENST00000557157 | HNRNPC-229 | 698 | - | ENSP00000450601 | 214 (aa) | - | G3V2D6 |
| ENST00000557033 | HNRNPC-228 | 760 | - | - | - (aa) | - | - |
| ENST00000556897 | HNRNPC-227 | 1252 | - | ENSP00000451176 | 293 (aa) | - | P07910 |
| ENST00000555585 | HNRNPC-220 | 682 | - | - | - (aa) | - | - |
| ENST00000553300 | HNRNPC-205 | 1375 | XM_006720125 | ENSP00000450544 | 293 (aa) | XP_006720188 | P07910 |
| ENST00000556226 | HNRNPC-224 | 629 | - | ENSP00000451292 | 121 (aa) | - | G3V3K6 |
| ENST00000554539 | HNRNPC-212 | 748 | - | ENSP00000452545 | 180 (aa) | - | G3V5V7 |
| ENST00000555137 | HNRNPC-216 | 789 | - | ENSP00000452185 | 175 (aa) | - | G3V555 |
| ENST00000555176 | HNRNPC-217 | 531 | - | ENSP00000452573 | 117 (aa) | - | G3V5X6 |
| ENST00000554891 | HNRNPC-213 | 644 | - | ENSP00000450467 | 126 (aa) | - | G3V251 |
| ENST00000557336 | HNRNPC-231 | 580 | - | ENSP00000451892 | 66 (aa) | - | G3V4M8 |
| ENST00000554383 | HNRNPC-209 | 1039 | - | ENSP00000452021 | 262 (aa) | - | G3V4W0 |
| ENST00000554455 | HNRNPC-211 | 1784 | XM_011536708 | ENSP00000451291 | 306 (aa) | XP_011535010 | P07910 |
| ENST00000555309 | HNRNPC-219 | 1714 | - | ENSP00000450790 | 305 (aa) | - | G3V2Q1 |
| ENST00000554417 | HNRNPC-210 | 691 | - | - | - (aa) | - | - |
| ENST00000556513 | HNRNPC-225 | 1403 | - | ENSP00000452214 | 231 (aa) | - | G3V576 |
| ENST00000556142 | HNRNPC-223 | 1599 | - | ENSP00000451187 | 231 (aa) | - | G3V576 |
| ENST00000420743 | HNRNPC-202 | 1559 | XM_024449556 | ENSP00000404848 | 306 (aa) | XP_024305324 | P07910 |
| ENST00000555883 | HNRNPC-221 | 1182 | - | ENSP00000450629 | 250 (aa) | - | P07910 |
| ENST00000553753 | HNRNPC-208 | 1790 | - | ENSP00000450548 | 288 (aa) | - | B4DY08 |
| ENST00000553444 | HNRNPC-206 | 724 | - | - | - (aa) | - | - |
| ENST00000555914 | HNRNPC-222 | 1658 | - | ENSP00000451708 | 292 (aa) | - | G3V4C1 |
| ENST00000449098 | HNRNPC-204 | 1599 | - | ENSP00000404559 | 290 (aa) | - | B2R5W2 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03040 | Spliceosome | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000092199 | rs17197037 | 14 | 21257495 | A | Bipolar disorder | 21738484 | [NR] | 1.17 | EFO_0000289 |
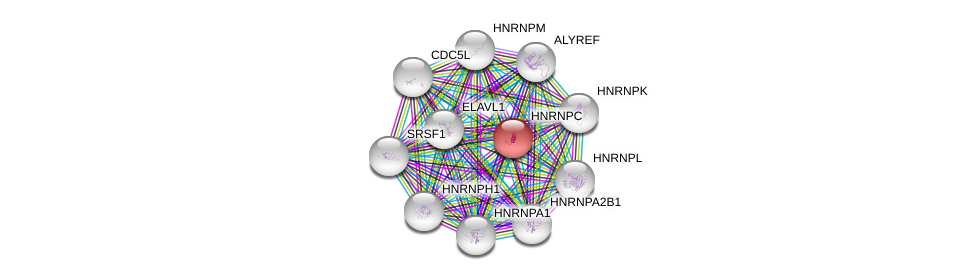
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000092199 | HNRNPC | 62 | 32.500 | ENSG00000213516 | RBMXL1 | 55 | 32.955 |
| ENSG00000092199 | HNRNPC | 76 | 42.000 | ENSG00000100650 | SRSF5 | 82 | 35.366 |
| ENSG00000092199 | HNRNPC | 100 | 94.737 | ENSG00000179172 | HNRNPCL1 | 100 | 90.785 |
| ENSG00000092199 | HNRNPC | 100 | 80.303 | ENSG00000125970 | RALY | 88 | 83.784 |
| ENSG00000092199 | HNRNPC | 62 | 34.667 | ENSG00000115875 | SRSF7 | 56 | 34.667 |
| ENSG00000092199 | HNRNPC | 96 | 84.615 | ENSG00000184672 | RALYL | 92 | 80.769 |
| ENSG00000092199 | HNRNPC | 52 | 30.263 | ENSG00000124193 | SRSF6 | 56 | 30.263 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSAMEG00000006492 | HNRNPC | 87 | 94.667 | Ailuropoda_melanoleuca |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSACAG00000029093 | - | 97 | 90.523 | Anolis_carolinensis |
| ENSG00000092199 | HNRNPC | 100 | 98.473 | ENSANAG00000038269 | - | 100 | 96.013 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 93.939 | ENSANAG00000029835 | - | 100 | 88.406 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 93 | 98.000 | ENSANAG00000034055 | - | 97 | 89.423 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSANAG00000034906 | HNRNPC | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 88.889 | ENSANAG00000025823 | - | 84 | 86.312 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 96 | 94.220 | ENSANAG00000037534 | - | 98 | 89.952 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 96 | 93.750 | ENSANAG00000018389 | - | 100 | 79.452 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 92.188 | ENSANAG00000031628 | - | 100 | 91.186 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 90.741 | ENSANAG00000027016 | - | 97 | 87.739 | Aotus_nancymaae |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSBTAG00000011396 | HNRNPC | 100 | 98.299 | Bos_taurus |
| ENSG00000092199 | HNRNPC | 100 | 80.303 | ENSBTAG00000049420 | - | 95 | 86.735 | Bos_taurus |
| ENSG00000092199 | HNRNPC | 100 | 98.092 | ENSCJAG00000023213 | - | 97 | 96.121 | Callithrix_jacchus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCJAG00000003238 | - | 94 | 99.257 | Callithrix_jacchus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCJAG00000016319 | - | 99 | 93.728 | Callithrix_jacchus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCJAG00000009354 | HNRNPC | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCAFG00000002619 | - | 100 | 94.539 | Canis_familiaris |
| ENSG00000092199 | HNRNPC | 100 | 98.413 | ENSCAFG00000018380 | - | 100 | 96.939 | Canis_familiaris |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCAFG00000005504 | - | 90 | 94.667 | Canis_familiaris |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCAFG00020014469 | HNRNPC | 95 | 98.127 | Canis_lupus_dingo |
| ENSG00000092199 | HNRNPC | 100 | 92.818 | ENSCHIG00000026850 | - | 100 | 88.104 | Capra_hircus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCHIG00000022536 | - | 95 | 98.127 | Capra_hircus |
| ENSG00000092199 | HNRNPC | 100 | 86.364 | ENSCHIG00000015259 | - | 100 | 62.500 | Capra_hircus |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSTSYG00000034314 | - | 78 | 79.147 | Carlito_syrichta |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSTSYG00000003305 | - | 100 | 97.854 | Carlito_syrichta |
| ENSG00000092199 | HNRNPC | 100 | 85.185 | ENSTSYG00000026064 | - | 88 | 82.534 | Carlito_syrichta |
| ENSG00000092199 | HNRNPC | 99 | 94.828 | ENSTSYG00000027744 | - | 100 | 80.272 | Carlito_syrichta |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCAPG00000008726 | HNRNPC | 100 | 95.192 | Cavia_aperea |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCPOG00000001474 | HNRNPC | 100 | 97.068 | Cavia_porcellus |
| ENSG00000092199 | HNRNPC | 76 | 89.888 | ENSCCAG00000014874 | - | 97 | 79.237 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCCAG00000035141 | - | 87 | 100.000 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSCCAG00000020533 | - | 93 | 96.992 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 80 | 87.129 | ENSCCAG00000023170 | - | 100 | 71.053 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 100 | 97.619 | ENSCCAG00000029854 | - | 100 | 93.515 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCCAG00000025125 | - | 100 | 100.000 | Cebus_capucinus |
| ENSG00000092199 | HNRNPC | 82 | 100.000 | ENSCATG00000030828 | - | 92 | 95.536 | Cercocebus_atys |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCATG00000035936 | - | 93 | 99.624 | Cercocebus_atys |
| ENSG00000092199 | HNRNPC | 100 | 95.556 | ENSCATG00000031526 | - | 100 | 93.857 | Cercocebus_atys |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCLAG00000000947 | HNRNPC | 100 | 97.720 | Chinchilla_lanigera |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCSAG00000016585 | HNRNPC | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000092199 | HNRNPC | 100 | 94.737 | ENSCSAG00000000637 | - | 100 | 80.205 | Chlorocebus_sabaeus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCHOG00000010033 | HNRNPC | 100 | 78.495 | Choloepus_hoffmanni |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCPBG00000011437 | - | 98 | 90.942 | Chrysemys_picta_bellii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCANG00000035615 | HNRNPC | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSCANG00000037885 | - | 88 | 90.421 | Colobus_angolensis_palliatus |
| ENSG00000092199 | HNRNPC | 99 | 91.188 | ENSCANG00000034732 | - | 98 | 90.875 | Colobus_angolensis_palliatus |
| ENSG00000092199 | HNRNPC | 100 | 81.481 | ENSCANG00000031846 | - | 85 | 73.551 | Colobus_angolensis_palliatus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCGRG00001004415 | Hnrnpc | 100 | 95.860 | Cricetulus_griseus_chok1gshd |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSCGRG00001006054 | - | 100 | 68.198 | Cricetulus_griseus_chok1gshd |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSCGRG00001024282 | - | 97 | 81.679 | Cricetulus_griseus_chok1gshd |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSCGRG00000012098 | Hnrnpc | 100 | 98.046 | Cricetulus_griseus_crigri |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSCGRG00000009850 | - | 97 | 81.679 | Cricetulus_griseus_crigri |
| ENSG00000092199 | HNRNPC | 90 | 75.221 | ENSDNOG00000043205 | - | 97 | 64.865 | Dasypus_novemcinctus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSDNOG00000040064 | - | 100 | 96.732 | Dasypus_novemcinctus |
| ENSG00000092199 | HNRNPC | 100 | 94.872 | ENSDNOG00000044551 | - | 100 | 90.411 | Dasypus_novemcinctus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSDNOG00000003161 | - | 100 | 98.371 | Dasypus_novemcinctus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSDORG00000012163 | - | 100 | 86.054 | Dipodomys_ordii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSEASG00005010413 | HNRNPC | 100 | 98.697 | Equus_asinus_asinus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSECAG00000017645 | HNRNPC | 100 | 98.697 | Equus_caballus |
| ENSG00000092199 | HNRNPC | 100 | 98.857 | ENSEEUG00000007472 | HNRNPC | 100 | 95.455 | Erinaceus_europaeus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSFCAG00000014889 | HNRNPC | 100 | 98.046 | Felis_catus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSFDAG00000003406 | - | 100 | 97.720 | Fukomys_damarensis |
| ENSG00000092199 | HNRNPC | 100 | 96.970 | ENSFDAG00000010728 | - | 99 | 87.500 | Fukomys_damarensis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSGAGG00000011310 | - | 98 | 89.493 | Gopherus_agassizii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSGGOG00000023015 | HNRNPC | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSGGOG00000022340 | - | 94 | 85.433 | Gorilla_gorilla |
| ENSG00000092199 | HNRNPC | 100 | 77.222 | ENSGGOG00000028510 | - | 100 | 70.805 | Gorilla_gorilla |
| ENSG00000092199 | HNRNPC | 100 | 84.127 | ENSHGLG00000006562 | - | 99 | 78.082 | Heterocephalus_glaber_female |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSHGLG00000014317 | - | 97 | 78.868 | Heterocephalus_glaber_female |
| ENSG00000092199 | HNRNPC | 100 | 92.562 | ENSHGLG00000015984 | - | 100 | 69.966 | Heterocephalus_glaber_female |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSHGLG00000005772 | HNRNPC | 100 | 97.394 | Heterocephalus_glaber_female |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSHGLG00100001282 | - | 100 | 96.743 | Heterocephalus_glaber_male |
| ENSG00000092199 | HNRNPC | 100 | 92.562 | ENSHGLG00100017195 | - | 100 | 69.966 | Heterocephalus_glaber_male |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSHGLG00100019072 | - | 97 | 78.868 | Heterocephalus_glaber_male |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSSTOG00000030323 | - | 91 | 98.667 | Ictidomys_tridecemlineatus |
| ENSG00000092199 | HNRNPC | 98 | 86.154 | ENSSTOG00000021413 | - | 93 | 73.856 | Ictidomys_tridecemlineatus |
| ENSG00000092199 | HNRNPC | 100 | 98.148 | ENSJJAG00000000615 | - | 95 | 76.336 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 100 | 96.970 | ENSJJAG00000017581 | - | 100 | 84.642 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 100 | 93.939 | ENSJJAG00000019199 | - | 93 | 89.354 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 99 | 100.000 | ENSJJAG00000014312 | - | 100 | 94.788 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 99 | 100.000 | ENSJJAG00000019720 | - | 96 | 82.295 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 100 | 95.455 | ENSJJAG00000020950 | - | 100 | 92.708 | Jaculus_jaculus |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSLACG00000012197 | - | 97 | 76.846 | Latimeria_chalumnae |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSLAFG00000009341 | HNRNPC | 89 | 95.623 | Loxodonta_africana |
| ENSG00000092199 | HNRNPC | 100 | 90.741 | ENSMFAG00000002431 | - | 97 | 82.243 | Macaca_fascicularis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMFAG00000000371 | - | 95 | 98.876 | Macaca_fascicularis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMFAG00000002531 | HNRNPC | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000092199 | HNRNPC | 94 | 96.450 | ENSMFAG00000031240 | - | 95 | 97.059 | Macaca_fascicularis |
| ENSG00000092199 | HNRNPC | 100 | 87.037 | ENSMMUG00000016162 | - | 89 | 66.667 | Macaca_mulatta |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMMUG00000014809 | HNRNPC | 100 | 100.000 | Macaca_mulatta |
| ENSG00000092199 | HNRNPC | 100 | 98.333 | ENSMMUG00000018813 | - | 100 | 98.069 | Macaca_mulatta |
| ENSG00000092199 | HNRNPC | 100 | 94.737 | ENSMMUG00000046109 | - | 98 | 80.612 | Macaca_mulatta |
| ENSG00000092199 | HNRNPC | 100 | 98.148 | ENSMMUG00000001089 | - | 90 | 96.629 | Macaca_mulatta |
| ENSG00000092199 | HNRNPC | 100 | 74.242 | ENSMNEG00000041647 | - | 97 | 64.151 | Macaca_nemestrina |
| ENSG00000092199 | HNRNPC | 100 | 99.600 | ENSMNEG00000042280 | - | 100 | 99.600 | Macaca_nemestrina |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMLEG00000040545 | - | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000092199 | HNRNPC | 100 | 95.556 | ENSMLEG00000036782 | - | 100 | 94.198 | Mandrillus_leucophaeus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMAUG00000000739 | Hnrnpc | 100 | 98.046 | Mesocricetus_auratus |
| ENSG00000092199 | HNRNPC | 100 | 80.597 | ENSMAUG00000020887 | - | 97 | 72.263 | Mesocricetus_auratus |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSMICG00000031852 | - | 99 | 85.902 | Microcebus_murinus |
| ENSG00000092199 | HNRNPC | 100 | 90.741 | ENSMICG00000028823 | - | 87 | 78.261 | Microcebus_murinus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMICG00000007682 | HNRNPC | 100 | 99.320 | Microcebus_murinus |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSMOCG00000001623 | - | 100 | 95.349 | Microtus_ochrogaster |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMOCG00000020993 | - | 100 | 85.106 | Microtus_ochrogaster |
| ENSG00000092199 | HNRNPC | 100 | 85.185 | ENSMODG00000017619 | - | 97 | 77.033 | Monodelphis_domestica |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSMODG00000006606 | HNRNPC | 100 | 97.959 | Monodelphis_domestica |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMUSG00000060373 | Hnrnpc | 100 | 100.000 | Mus_musculus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | MGP_PahariEiJ_G0030256 | Hnrnpc | 100 | 93.312 | Mus_pahari |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | MGP_SPRETEiJ_G0020126 | Hnrnpc | 100 | 97.394 | Mus_spretus |
| ENSG00000092199 | HNRNPC | 100 | 98.148 | ENSMPUG00000016707 | - | 97 | 95.802 | Mustela_putorius_furo |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMPUG00000000164 | - | 100 | 97.952 | Mustela_putorius_furo |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSMLUG00000007853 | HNRNPC | 100 | 96.939 | Myotis_lucifugus |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSNGAG00000020552 | - | 100 | 93.333 | Nannospalax_galili |
| ENSG00000092199 | HNRNPC | 100 | 90.741 | ENSNGAG00000017956 | - | 100 | 71.480 | Nannospalax_galili |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSNGAG00000017860 | - | 99 | 77.133 | Nannospalax_galili |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSNLEG00000014362 | HNRNPC | 99 | 100.000 | Nomascus_leucogenys |
| ENSG00000092199 | HNRNPC | 100 | 96.296 | ENSNLEG00000018726 | - | 98 | 96.198 | Nomascus_leucogenys |
| ENSG00000092199 | HNRNPC | 100 | 96.296 | ENSNLEG00000009500 | - | 100 | 75.085 | Nomascus_leucogenys |
| ENSG00000092199 | HNRNPC | 100 | 87.037 | ENSNLEG00000028034 | - | 85 | 71.377 | Nomascus_leucogenys |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSMEUG00000010928 | - | 64 | 98.305 | Notamacropus_eugenii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOPRG00000002800 | HNRNPC | 82 | 100.000 | Ochotona_princeps |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSODEG00000001160 | - | 98 | 90.345 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 100 | 85.165 | ENSODEG00000017188 | - | 100 | 82.034 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 98 | 84.906 | ENSODEG00000012803 | - | 100 | 79.739 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSODEG00000000603 | - | 100 | 93.312 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSODEG00000008102 | - | 100 | 95.578 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSODEG00000020161 | - | 100 | 93.811 | Octodon_degus |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSOANG00000015707 | HNRNPC | 100 | 97.959 | Ornithorhynchus_anatinus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOCUG00000000339 | - | 100 | 98.046 | Oryctolagus_cuniculus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOCUG00000013026 | - | 97 | 90.909 | Oryctolagus_cuniculus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOCUG00000015029 | HNRNPC | 100 | 98.046 | Oryctolagus_cuniculus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSOCUG00000008851 | - | 100 | 97.068 | Oryctolagus_cuniculus |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSOGAG00000029866 | - | 100 | 89.153 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOGAG00000033117 | - | 100 | 96.124 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 88.889 | ENSOGAG00000032637 | - | 96 | 74.886 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 88.889 | ENSOGAG00000032179 | - | 100 | 76.144 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOGAG00000028828 | - | 100 | 89.865 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOGAG00000012543 | - | 100 | 97.288 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOGAG00000032867 | - | 100 | 96.271 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOGAG00000032454 | - | 100 | 94.576 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 86.364 | ENSOGAG00000034058 | - | 97 | 58.842 | Otolemur_garnettii |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSOARG00000010292 | - | 97 | 93.333 | Ovis_aries |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSOARG00000019672 | - | 99 | 94.463 | Ovis_aries |
| ENSG00000092199 | HNRNPC | 100 | 80.000 | ENSPPAG00000033436 | - | 100 | 73.154 | Pan_paniscus |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSPPAG00000041389 | - | 98 | 88.517 | Pan_paniscus |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSPPAG00000030712 | - | 95 | 81.690 | Pan_paniscus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPPAG00000034008 | - | 100 | 100.000 | Pan_paniscus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPPRG00000008763 | HNRNPC | 100 | 98.046 | Panthera_pardus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPTIG00000021061 | HNRNPC | 100 | 98.299 | Panthera_tigris_altaica |
| ENSG00000092199 | HNRNPC | 100 | 81.111 | ENSPTRG00000045559 | - | 100 | 72.819 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 88 | 88.608 | ENSPTRG00000043617 | - | 87 | 89.542 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPTRG00000006118 | HNRNPC | 100 | 100.000 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSPTRG00000043496 | - | 86 | 86.977 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 100 | 88.889 | ENSPTRG00000039710 | - | 99 | 88.785 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 100 | 92.593 | ENSPTRG00000043469 | - | 100 | 80.137 | Pan_troglodytes |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPANG00000031574 | - | 86 | 100.000 | Papio_anubis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPANG00000020782 | - | 100 | 100.000 | Papio_anubis |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSPANG00000035897 | - | 100 | 94.881 | Papio_anubis |
| ENSG00000092199 | HNRNPC | 92 | 97.576 | ENSPANG00000018274 | - | 94 | 97.436 | Papio_anubis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPSIG00000012177 | - | 100 | 92.157 | Pelodiscus_sinensis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPEMG00000017899 | - | 100 | 98.046 | Peromyscus_maniculatus_bairdii |
| ENSG00000092199 | HNRNPC | 100 | 94.444 | ENSPEMG00000017675 | - | 100 | 75.503 | Peromyscus_maniculatus_bairdii |
| ENSG00000092199 | HNRNPC | 100 | 99.206 | ENSPCIG00000020021 | - | 100 | 97.952 | Phascolarctos_cinereus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPPYG00000005593 | HNRNPC | 100 | 100.000 | Pongo_abelii |
| ENSG00000092199 | HNRNPC | 100 | 94.737 | ENSPPYG00000001860 | - | 100 | 89.420 | Pongo_abelii |
| ENSG00000092199 | HNRNPC | 100 | 96.970 | ENSPCAG00000003025 | HNRNPC | 100 | 80.392 | Procavia_capensis |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPCOG00000022866 | HNRNPC | 100 | 99.320 | Propithecus_coquereli |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSPVAG00000004499 | HNRNPC | 91 | 97.857 | Pteropus_vampyrus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSRNOG00000011621 | LOC100911576 | 100 | 97.619 | Rattus_norvegicus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSRNOG00000050800 | Hnrnpc | 100 | 95.349 | Rattus_norvegicus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSRBIG00000038355 | HNRNPC | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000092199 | HNRNPC | 100 | 97.368 | ENSRBIG00000006622 | - | 98 | 80.887 | Rhinopithecus_bieti |
| ENSG00000092199 | HNRNPC | 94 | 77.143 | ENSRROG00000029991 | - | 84 | 69.925 | Rhinopithecus_roxellana |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSRROG00000036842 | - | 100 | 96.654 | Rhinopithecus_roxellana |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSRROG00000031669 | - | 100 | 95.257 | Rhinopithecus_roxellana |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSRROG00000006722 | HNRNPC | 100 | 92.675 | Rhinopithecus_roxellana |
| ENSG00000092199 | HNRNPC | 100 | 97.521 | ENSSBOG00000030368 | - | 82 | 79.012 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 88 | 87.446 | ENSSBOG00000028896 | - | 97 | 87.295 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 99 | 92.105 | ENSSBOG00000022725 | - | 99 | 78.082 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 100 | 99.237 | ENSSBOG00000024757 | HNRNPC | 100 | 99.134 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 99 | 81.897 | ENSSBOG00000032742 | - | 100 | 70.428 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 100 | 94.017 | ENSSBOG00000028154 | - | 98 | 85.057 | Saimiri_boliviensis_boliviensis |
| ENSG00000092199 | HNRNPC | 100 | 98.857 | ENSSHAG00000006145 | HNRNPC | 100 | 96.452 | Sarcophilus_harrisii |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSSARG00000003711 | HNRNPC | 100 | 79.085 | Sorex_araneus |
| ENSG00000092199 | HNRNPC | 100 | 98.485 | ENSSPUG00000018728 | - | 100 | 90.523 | Sphenodon_punctatus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSSSCG00000038947 | HNRNPC | 100 | 98.639 | Sus_scrofa |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSTBEG00000011514 | - | 100 | 89.362 | Tupaia_belangeri |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSTTRG00000016055 | HNRNPC | 100 | 98.371 | Tursiops_truncatus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSUMAG00000003799 | HNRNPC | 100 | 98.039 | Ursus_maritimus |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSVPAG00000007337 | HNRNPC | 100 | 98.046 | Vicugna_pacos |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSVVUG00000009265 | - | 95 | 98.127 | Vulpes_vulpes |
| ENSG00000092199 | HNRNPC | 100 | 100.000 | ENSVVUG00000012230 | - | 90 | 97.826 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | 9731529.11991638. | IC | Process |
| GO:0000398 | mRNA splicing, via spliceosome | 25719671. | IMP | Process |
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0000790 | nuclear chromatin | 16217013. | HDA | Component |
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 16217013. | HDA | Function |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 16217013. | HDA | Function |
| GO:0001649 | osteoblast differentiation | 16210410. | HDA | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 18082603. | IPI | Function |
| GO:0003723 | RNA binding | 9731529. | NAS | Function |
| GO:0003730 | mRNA 3'-UTR binding | 16010978. | IDA | Function |
| GO:0005515 | protein binding | 11546873.11687588.15034300.15161933.16189514.17289661.19447967.20467437.20618440.21516116.21988832.22365833.22446626.22720776.23602568.24965446.25416956.30021884. | IPI | Function |
| GO:0005576 | extracellular region | 27068509. | HDA | Component |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 11687588.22720776. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005681 | spliceosomal complex | 9731529. | IDA | Component |
| GO:0005697 | telomerase holoenzyme complex | 18082603. | IDA | Component |
| GO:0005829 | cytosol | 16010978. | IDA | Component |
| GO:0008266 | poly(U) RNA binding | 16010978. | IDA | Function |
| GO:0008380 | RNA splicing | 3110598. | TAS | Process |
| GO:0015629 | actin cytoskeleton | 11687588. | IDA | Component |
| GO:0016020 | membrane | 16210410. | HDA | Component |
| GO:0016070 | RNA metabolic process | - | TAS | Process |
| GO:0031492 | nucleosomal DNA binding | 16217013. | HDA | Function |
| GO:0032211 | negative regulation of telomere maintenance via telomerase | 18082603. | IMP | Process |
| GO:0032991 | protein-containing complex | 16217013. | HDA | Component |
| GO:0032991 | protein-containing complex | 11687588. | IDA | Component |
| GO:0042802 | identical protein binding | 16189514.21516116.22365833.25416956. | IPI | Function |
| GO:0043044 | ATP-dependent chromatin remodeling | 16217013. | HDA | Process |
| GO:0070034 | telomerase RNA binding | 18082603. | IPI | Function |
| GO:0070062 | extracellular exosome | 20458337. | HDA | Component |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 16010978. | IMP | Process |
| GO:0071013 | catalytic step 2 spliceosome | 11991638. | IDA | Component |
| GO:1990247 | N6-methyladenosine-containing RNA binding | 25719671. | IDA | Function |