
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27164557 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000405486 | RANGAP1-202 | 3469 | XM_005261695 | ENSP00000385866 | 587 (aa) | XP_005261752 | P46060 |
| ENST00000446258 | RANGAP1-205 | 759 | - | ENSP00000393407 | 253 (aa) | - | H0Y4Q3 |
| ENST00000356244 | RANGAP1-201 | 3060 | XM_006724289 | ENSP00000348577 | 587 (aa) | XP_006724352 | P46060 |
| ENST00000455915 | RANGAP1-207 | 4222 | XM_005261696 | ENSP00000401470 | 587 (aa) | XP_005261753 | P46060 |
| ENST00000418067 | RANGAP1-203 | 650 | - | ENSP00000408743 | 100 (aa) | - | B0QYT5 |
| ENST00000452543 | RANGAP1-206 | 603 | - | ENSP00000394787 | 133 (aa) | - | B0QYT4 |
| ENST00000422838 | RANGAP1-204 | 946 | - | ENSP00000404842 | 37 (aa) | - | B0QYT6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03013 | RNA transport | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000100401 | rs2235852 | 22 | 41265150 | T | Self-reported allergy | 23817569 | [0.033-0.082] unit increase | 0.0577 | EFO_0003785 |
| ENSG00000100401 | rs3171692 | 22 | 41246208 | T | Feeling tense | 29500382 | z score increase | 5.66 | EFO_0009596 |
| ENSG00000100401 | rs139498 | 22 | 41248230 | C | Adventurousness | 30643258 | [0.009-0.0167] unit increase | 0.012839883 | EFO_0008579 |
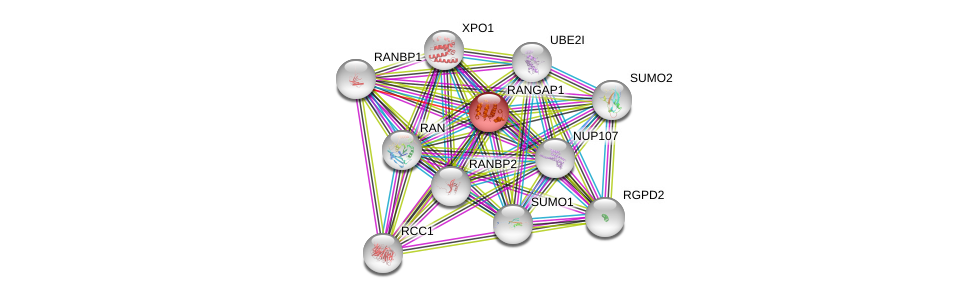
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100401 | RANGAP1 | 100 | 45.113 | FBgn0003346 | RanGAP | 57 | 44.575 | Drosophila_melanogaster |
| ENSG00000100401 | RANGAP1 | 100 | 97.297 | ENSMUSG00000022391 | Rangap1 | 100 | 95.833 | Mus_musculus |
| ENSG00000100401 | RANGAP1 | 83 | 32.456 | YMR235C | 79 | 30.675 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000776 | kinetochore | 17363900. | IDA | Component |
| GO:0000777 | condensed chromosome kinetochore | - | IEA | Component |
| GO:0003723 | RNA binding | 26308891. | IDA | Function |
| GO:0005096 | GTPase activator activity | 16428860. | IDA | Function |
| GO:0005515 | protein binding | 14729961.16732283.17000644.17036045.17099698.17099700.18093978.19549727.20414307.21988832.23395904.26496610. | IPI | Function |
| GO:0005635 | nuclear envelope | 25468996. | IDA | Component |
| GO:0005635 | nuclear envelope | - | TAS | Component |
| GO:0005643 | nuclear pore | 8978815. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IEA | Component |
| GO:0005737 | cytoplasm | 8978815. | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007165 | signal transduction | - | IEA | Process |
| GO:0008536 | Ran GTPase binding | 16428860. | IPI | Function |
| GO:0016235 | aggresome | - | IDA | Component |
| GO:0016925 | protein sumoylation | - | TAS | Process |
| GO:0030425 | dendrite | - | IEA | Component |
| GO:0031625 | ubiquitin protein ligase binding | - | IEA | Function |
| GO:0031965 | nuclear membrane | - | IDA | Component |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0044614 | nuclear pore cytoplasmic filaments | - | IEA | Component |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0046826 | negative regulation of protein export from nucleus | 16449645. | IDA | Process |
| GO:0048471 | perinuclear region of cytoplasm | 26308891. | IDA | Component |
| GO:0048678 | response to axon injury | - | IEA | Process |
| GO:0072686 | mitotic spindle | - | IEA | Component |
| GO:0090630 | activation of GTPase activity | 16428860. | IDA | Process |
| GO:1904115 | axon cytoplasm | - | IEA | Component |
| GO:1904117 | cellular response to vasopressin | - | IEA | Process |
| GO:1990723 | cytoplasmic periphery of the nuclear pore complex | - | IEA | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| PCPG | |||
| SKCM | |||
| THCA | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| ACC | ||
| SKCM | ||
| KIRP | ||
| PCPG | ||
| CESC | ||
| READ | ||
| LAML | ||
| LIHC | ||
| DLBC | ||
| LUAD |