
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000219782 | zf-C2H2 | PF00096.26 | 2.8e-25 | 1 | 2 |
| ENSP00000219782 | zf-C2H2 | PF00096.26 | 2.8e-25 | 2 | 2 |
| ENSP00000443956 | zf-C2H2 | PF00096.26 | 1.6e-19 | 1 | 2 |
| ENSP00000443956 | zf-C2H2 | PF00096.26 | 1.6e-19 | 2 | 2 |
| ENSP00000313362 | zf-C2H2 | PF00096.26 | 2.4e-19 | 1 | 2 |
| ENSP00000313362 | zf-C2H2 | PF00096.26 | 2.4e-19 | 2 | 2 |
| ENSP00000460057 | zf-C2H2 | PF00096.26 | 1e-17 | 1 | 4 |
| ENSP00000460057 | zf-C2H2 | PF00096.26 | 1e-17 | 2 | 4 |
| ENSP00000460057 | zf-C2H2 | PF00096.26 | 1e-17 | 3 | 4 |
| ENSP00000460057 | zf-C2H2 | PF00096.26 | 1e-17 | 4 | 4 |
| ENSP00000480440 | zf-C2H2 | PF00096.26 | 1.1e-17 | 1 | 3 |
| ENSP00000480440 | zf-C2H2 | PF00096.26 | 1.1e-17 | 2 | 3 |
| ENSP00000480440 | zf-C2H2 | PF00096.26 | 1.1e-17 | 3 | 3 |
| ENSP00000455794 | zf-C2H2 | PF00096.26 | 7.3e-11 | 1 | 2 |
| ENSP00000455794 | zf-C2H2 | PF00096.26 | 7.3e-11 | 2 | 2 |
| ENSP00000461630 | zf-C2H2 | PF00096.26 | 9.9e-06 | 1 | 1 |
| ENSP00000458201 | zf-C2H2 | PF00096.26 | 2.3e-05 | 1 | 1 |
| ENSP00000219782 | zf-met | PF12874.7 | 3.1e-08 | 1 | 1 |
| ENSP00000458201 | zf-met | PF12874.7 | 2.7e-07 | 1 | 2 |
| ENSP00000458201 | zf-met | PF12874.7 | 2.7e-07 | 2 | 2 |
| ENSP00000461630 | zf-met | PF12874.7 | 2.1e-06 | 1 | 2 |
| ENSP00000461630 | zf-met | PF12874.7 | 2.1e-06 | 2 | 2 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 22198906 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000616501 | MAZ-216 | 991 | - | ENSP00000480440 | 252 (aa) | - | A0A087WWR2 |
| ENST00000568544 | MAZ-214 | 1464 | - | ENSP00000455473 | 78 (aa) | - | H3BPU3 |
| ENST00000568411 | MAZ-213 | 599 | - | ENSP00000455794 | 196 (aa) | - | H3BQI4 |
| ENST00000545521 | MAZ-203 | 2333 | - | ENSP00000443956 | 454 (aa) | - | P56270 |
| ENST00000322945 | MAZ-202 | 2532 | - | ENSP00000313362 | 477 (aa) | - | P56270 |
| ENST00000567444 | MAZ-211 | 635 | - | ENSP00000460057 | 202 (aa) | - | I3L2Z5 |
| ENST00000562557 | MAZ-206 | 475 | - | ENSP00000461630 | 78 (aa) | - | I3L4Y2 |
| ENST00000561855 | MAZ-204 | 672 | - | ENSP00000456179 | 84 (aa) | - | H3BRC5 |
| ENST00000568282 | MAZ-212 | 1593 | - | ENSP00000458201 | 94 (aa) | - | I3L0M3 |
| ENST00000562337 | MAZ-205 | 1574 | - | ENSP00000455726 | 172 (aa) | - | P56270 |
| ENST00000563012 | MAZ-207 | 646 | - | ENSP00000455913 | 152 (aa) | - | H3BQS2 |
| ENST00000219782 | MAZ-201 | 2698 | XM_006721047 | ENSP00000219782 | 493 (aa) | XP_006721110 | P56270 |
| ENST00000566906 | MAZ-210 | 1455 | - | ENSP00000461174 | 184 (aa) | - | I3L4D3 |
| ENST00000565777 | MAZ-209 | 1313 | - | - | - (aa) | - | - |
| ENST00000563402 | MAZ-208 | 1461 | - | ENSP00000457310 | 186 (aa) | - | H3BTS8 |
| ENST00000569978 | MAZ-215 | 567 | - | ENSP00000460877 | 53 (aa) | - | I3L411 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000103495 | Multiple Sclerosis | 5E-7 | 27386562 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000103495 | rs34286592 | 16 | 29809159 | T | Multiple sclerosis | 27386562 | [1.09-1.23] | 1.16 | EFO_0003885 |
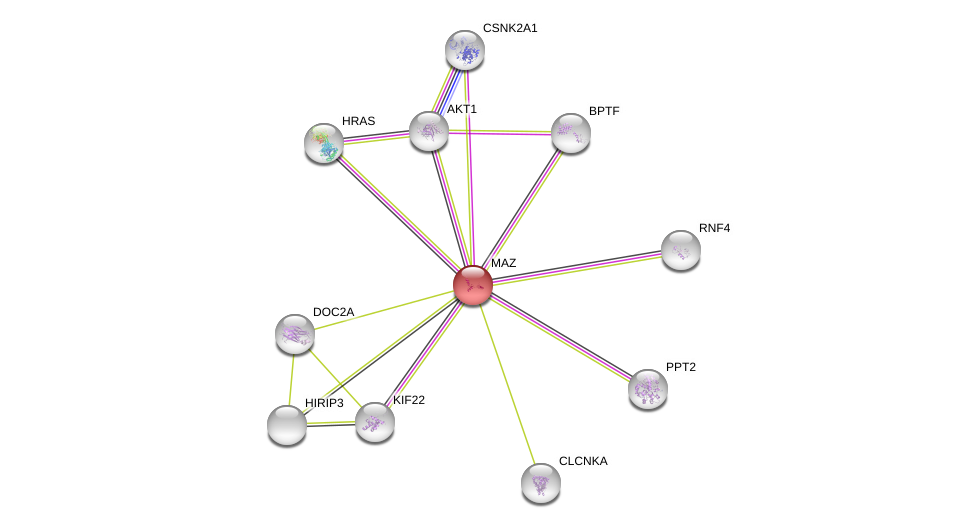
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000103495 | MAZ | 80 | 38.298 | ENSG00000169957 | ZNF768 | 57 | 37.662 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000103495 | MAZ | 100 | 85.897 | ENSANAG00000037536 | - | 100 | 88.542 | Aotus_nancymaae |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSANAG00000036195 | - | 100 | 93.927 | Aotus_nancymaae |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSBTAG00000052323 | MAZ | 100 | 99.394 | Bos_taurus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCJAG00000007189 | MAZ | 100 | 99.798 | Callithrix_jacchus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCAFG00000016964 | MAZ | 100 | 97.902 | Canis_familiaris |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCAFG00020017751 | MAZ | 100 | 99.192 | Canis_lupus_dingo |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCHIG00000020711 | MAZ | 100 | 99.394 | Capra_hircus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSTSYG00000026702 | - | 100 | 100.000 | Carlito_syrichta |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCAPG00000006391 | - | 100 | 100.000 | Cavia_aperea |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCCAG00000037524 | MAZ | 100 | 100.000 | Cebus_capucinus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCATG00000033033 | MAZ | 100 | 100.000 | Cercocebus_atys |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCLAG00000010257 | - | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSCSAG00000006110 | MAZ | 100 | 99.628 | Chlorocebus_sabaeus |
| ENSG00000103495 | MAZ | 96 | 84.000 | ENSCPBG00000012768 | - | 93 | 74.444 | Chrysemys_picta_bellii |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSCANG00000035162 | MAZ | 100 | 99.621 | Colobus_angolensis_palliatus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCGRG00001008879 | Maz | 100 | 98.174 | Cricetulus_griseus_chok1gshd |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSCGRG00000014850 | - | 100 | 99.526 | Cricetulus_griseus_crigri |
| ENSG00000103495 | MAZ | 76 | 78.431 | ENSCVAG00000015597 | si:ch211-166g5.4 | 52 | 78.947 | Cyprinodon_variegatus |
| ENSG00000103495 | MAZ | 96 | 60.526 | ENSDARG00000063555 | si:ch211-166g5.4 | 86 | 77.778 | Danio_rerio |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSEASG00005013172 | MAZ | 100 | 98.990 | Equus_asinus_asinus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSECAG00000024341 | MAZ | 100 | 98.990 | Equus_caballus |
| ENSG00000103495 | MAZ | 76 | 77.778 | ENSELUG00000022514 | si:ch211-166g5.4 | 77 | 77.419 | Esox_lucius |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSFCAG00000014146 | MAZ | 100 | 99.192 | Felis_catus |
| ENSG00000103495 | MAZ | 71 | 98.507 | ENSFDAG00000010852 | - | 100 | 74.408 | Fukomys_damarensis |
| ENSG00000103495 | MAZ | 82 | 80.714 | ENSFHEG00000000918 | MAZ | 91 | 80.714 | Fundulus_heteroclitus |
| ENSG00000103495 | MAZ | 96 | 69.737 | ENSGALG00000049721 | - | 64 | 69.737 | Gallus_gallus |
| ENSG00000103495 | MAZ | 76 | 79.085 | ENSGAFG00000000293 | si:ch211-166g5.4 | 57 | 79.605 | Gambusia_affinis |
| ENSG00000103495 | MAZ | 82 | 72.989 | ENSGACG00000014840 | maza | 89 | 67.257 | Gasterosteus_aculeatus |
| ENSG00000103495 | MAZ | 96 | 84.000 | ENSGAGG00000008124 | MAZ | 97 | 81.538 | Gopherus_agassizii |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSGGOG00000042750 | - | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000103495 | MAZ | 100 | 97.436 | ENSHGLG00000011024 | - | 96 | 82.576 | Heterocephalus_glaber_female |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSHGLG00000018743 | - | 100 | 98.106 | Heterocephalus_glaber_female |
| ENSG00000103495 | MAZ | 94 | 85.526 | ENSHGLG00100017872 | - | 89 | 79.909 | Heterocephalus_glaber_male |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSSTOG00000027842 | MAZ | 100 | 99.242 | Ictidomys_tridecemlineatus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSJJAG00000020922 | - | 100 | 98.864 | Jaculus_jaculus |
| ENSG00000103495 | MAZ | 100 | 73.077 | ENSLACG00000010161 | MAZ | 79 | 72.964 | Latimeria_chalumnae |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSMFAG00000040127 | MAZ | 100 | 99.621 | Macaca_fascicularis |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSMMUG00000019826 | MAZ | 98 | 83.641 | Macaca_mulatta |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSMNEG00000037329 | MAZ | 100 | 99.621 | Macaca_nemestrina |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSMLEG00000036147 | - | 100 | 98.936 | Mandrillus_leucophaeus |
| ENSG00000103495 | MAZ | 76 | 79.739 | ENSMAMG00000018743 | - | 72 | 80.263 | Mastacembelus_armatus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSMAUG00000011903 | Maz | 100 | 96.901 | Mesocricetus_auratus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSMICG00000030392 | MAZ | 100 | 99.594 | Microcebus_murinus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSMOCG00000003611 | Maz | 100 | 97.912 | Microtus_ochrogaster |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSMUSG00000030678 | Maz | 100 | 98.377 | Mus_musculus |
| ENSG00000103495 | MAZ | 100 | 100.000 | MGP_PahariEiJ_G0013693 | Maz | 100 | 98.580 | Mus_pahari |
| ENSG00000103495 | MAZ | 100 | 93.590 | ENSMEUG00000009482 | MAZ | 99 | 76.171 | Notamacropus_eugenii |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSODEG00000014451 | MAZ | 100 | 99.242 | Octodon_degus |
| ENSG00000103495 | MAZ | 78 | 94.937 | ENSOANG00000004112 | - | 77 | 97.980 | Ornithorhynchus_anatinus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSOARG00000005520 | MAZ | 99 | 93.231 | Ovis_aries |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPPAG00000041571 | - | 76 | 97.561 | Pan_paniscus |
| ENSG00000103495 | MAZ | 87 | 100.000 | ENSPPRG00000011596 | - | 100 | 87.121 | Panthera_pardus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPPRG00000016750 | - | 100 | 94.929 | Panthera_pardus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPTIG00000007866 | - | 100 | 100.000 | Panthera_tigris_altaica |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPTRG00000007968 | MAZ | 100 | 99.594 | Pan_troglodytes |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSPANG00000016381 | - | 100 | 99.621 | Papio_anubis |
| ENSG00000103495 | MAZ | 78 | 79.114 | ENSPKIG00000018202 | - | 80 | 70.183 | Paramormyrops_kingsleyae |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPEMG00000008318 | - | 100 | 91.469 | Peromyscus_maniculatus_bairdii |
| ENSG00000103495 | MAZ | 62 | 60.000 | ENSPMAG00000004866 | - | 82 | 50.655 | Petromyzon_marinus |
| ENSG00000103495 | MAZ | 85 | 72.414 | ENSPFOG00000005397 | si:ch211-166g5.4 | 52 | 79.605 | Poecilia_formosa |
| ENSG00000103495 | MAZ | 76 | 80.132 | ENSPLAG00000017219 | si:ch211-166g5.4 | 77 | 80.667 | Poecilia_latipinna |
| ENSG00000103495 | MAZ | 76 | 80.132 | ENSPMEG00000017552 | si:ch211-166g5.4 | 77 | 80.667 | Poecilia_mexicana |
| ENSG00000103495 | MAZ | 86 | 71.676 | ENSPREG00000023113 | si:ch211-166g5.4 | 55 | 79.605 | Poecilia_reticulata |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPPYG00000007260 | - | 100 | 100.000 | Pongo_abelii |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPCOG00000022370 | MAZ | 100 | 99.594 | Propithecus_coquereli |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSPVAG00000017209 | MAZ | 79 | 100.000 | Pteropus_vampyrus |
| ENSG00000103495 | MAZ | 85 | 71.930 | ENSPNAG00000004042 | si:ch211-166g5.4 | 85 | 67.532 | Pygocentrus_nattereri |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSRBIG00000001823 | MAZ | 100 | 99.621 | Rhinopithecus_bieti |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSRROG00000026952 | MAZ | 100 | 99.621 | Rhinopithecus_roxellana |
| ENSG00000103495 | MAZ | 100 | 98.936 | ENSSBOG00000020791 | - | 100 | 99.621 | Saimiri_boliviensis_boliviensis |
| ENSG00000103495 | MAZ | 86 | 74.566 | ENSSFOG00015021699 | si:ch211-166g5.4 | 60 | 74.500 | Scleropages_formosus |
| ENSG00000103495 | MAZ | 99 | 84.416 | ENSSPUG00000010618 | MAZ | 100 | 69.604 | Sphenodon_punctatus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSSSCG00000025321 | MAZ | 98 | 99.349 | Sus_scrofa |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSTTRG00000005420 | MAZ | 82 | 96.923 | Tursiops_truncatus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSUAMG00000004622 | MAZ | 100 | 100.000 | Ursus_americanus |
| ENSG00000103495 | MAZ | 100 | 100.000 | ENSVVUG00000026097 | MAZ | 100 | 100.000 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 1502157.25487955. | IDA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 25487955. | IDA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | NAS | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 11527412. | IPI | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1502157. | IDA | Process |
| GO:0006369 | termination of RNA polymerase II transcription | 1502157. | IDA | Process |
| GO:0008284 | positive regulation of cell proliferation | 25487955. | IDA | Process |
| GO:0010628 | positive regulation of gene expression | 25487955. | IDA | Process |
| GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 25487955. | IDA | Process |
| GO:0030335 | positive regulation of cell migration | 25487955. | IDA | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 25487955. | IDA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0051897 | positive regulation of protein kinase B signaling | 25487955. | IDA | Process |
| GO:2001234 | negative regulation of apoptotic signaling pathway | 25487955. | IDA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CHOL | |||
| LUAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| BLCA | ||
| LIHC | ||
| THCA |