
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000456420 | MMR_HSR1 | PF01926.23 | 5e-07 | 1 | 1 |
| ENSP00000454381 | MMR_HSR1 | PF01926.23 | 5.2e-07 | 1 | 1 |
| ENSP00000456638 | MMR_HSR1 | PF01926.23 | 8.2e-07 | 1 | 1 |
| ENSP00000261890 | MMR_HSR1 | PF01926.23 | 1.1e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000569304 | RAB11A-208 | 347 | - | - | - (aa) | - | - |
| ENST00000261890 | RAB11A-201 | 4930 | - | ENSP00000261890 | 216 (aa) | - | P62491 |
| ENST00000566233 | RAB11A-205 | 588 | - | ENSP00000454381 | 155 (aa) | - | H3BMH2 |
| ENST00000568850 | RAB11A-207 | 551 | - | - | - (aa) | - | - |
| ENST00000569896 | RAB11A-209 | 1396 | - | ENSP00000456420 | 155 (aa) | - | P62491 |
| ENST00000565075 | RAB11A-204 | 711 | - | ENSP00000456638 | 198 (aa) | - | H3BSC1 |
| ENST00000564910 | RAB11A-203 | 978 | - | ENSP00000455567 | 146 (aa) | - | B4DQU5 |
| ENST00000567671 | RAB11A-206 | 635 | - | ENSP00000454673 | 81 (aa) | - | H3BN38 |
| ENST00000563580 | RAB11A-202 | 553 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000103769 | Platelet Function Tests | 2.3369300E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000103769 | rs7181649 | 15 | 65762011 | C | Mean corpuscular hemoglobin | 27863252 | [0.027-0.043] unit decrease | 0.0346963 | EFO_0004527 |
| ENSG00000103769 | rs2572207 | 15 | 65778355 | T | Mean corpuscular hemoglobin | 27863252 | [0.068-0.084] unit decrease | 0.07631472 | EFO_0004527 |
| ENSG00000103769 | rs11326570 | 15 | 65789510 | T | Red blood cell count | 27863252 | [0.027-0.043] unit increase | 0.03509589 | EFO_0004305 |
| ENSG00000103769 | rs2414884 | 15 | 65783534 | T | Mean corpuscular volume | 27863252 | [0.046-0.06] unit decrease | 0.05271687 | EFO_0004526 |
| ENSG00000103769 | rs11315539 | 15 | 65805720 | C | Mean corpuscular volume | 27863252 | [0.056-0.072] unit increase | 0.06372744 | EFO_0004526 |
| ENSG00000103769 | rs77559483 | 15 | 65740788 | A | Blood protein levels | 29875488 | [0.52-0.92] unit increase | 0.72 | EFO_0007937 |
| ENSG00000103769 | rs6494537 | 15 | 65759007 | C | Mean corpuscular hemoglobin | 20139978 | [0.053-0.103] unit increase | 0.078 | EFO_0004509|EFO_0004527 |
| ENSG00000103769 | rs2572207 | 15 | 65778355 | ? | Mean corpuscular hemoglobin | 28017375 | EFO_0004527 | ||
| ENSG00000103769 | rs2572207 | 15 | 65778355 | C | Red blood cell traits | 23222517 | [0.096-0.210] unit increase | 0.153 | EFO_0004526 |
| ENSG00000103769 | rs72741274 | 15 | 65770237 | T | Intelligence (MTAG) | 29326435 | [0.025-0.05] unit decrease | 0.037347373 | EFO_0004337 |
| ENSG00000103769 | rs11857609 | 15 | 65802932 | ? | Mean corpuscular volume | 29403010 | [0.035-0.053] unit increase novel | 0.04421 | EFO_0004526 |
| ENSG00000103769 | rs11857609 | 15 | 65802932 | ? | Mean corpuscular hemoglobin | 29403010 | [0.042-0.06] unit increase novel | 0.05134 | EFO_0004527 |
| ENSG00000103769 | rs11857609 | 15 | 65802932 | ? | Mean corpuscular hemoglobin concentration | 29403010 | [0.021-0.04] unit increase novel | 0.03042 | EFO_0004528 |
| ENSG00000103769 | rs557277320 | 15 | 65757263 | T | Hemoglobin concentration | 27863252 | [0.014-0.028] unit decrease | 0.02112256 | EFO_0004509 |
| ENSG00000103769 | rs12899850 | 15 | 65758961 | C | Body mass index | 30108127 | [NR] unit decrease | 0.014 | EFO_0004340 |
| ENSG00000103769 | rs61751117 | 15 | 65741731 | C | Logical memory (delayed recall) in normal cognition | 29274321 | [0.47-1.09] unit increase | 0.78 | EFO_0004874 |
| ENSG00000103769 | rs28498859 | 15 | 65775439 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000103769 | rs28498859 | 15 | 65775439 | ? | Red blood cell count | 30595370 | EFO_0004305 |
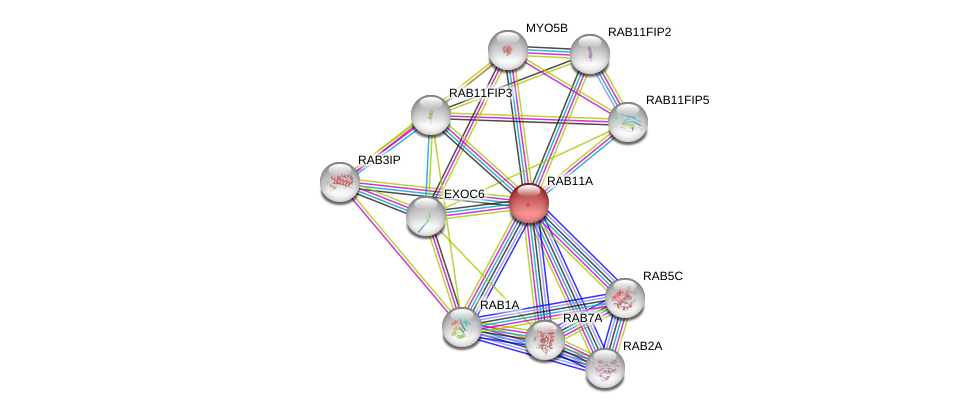
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSAPOG00000018256 | rab11a | 100 | 94.907 | Acanthochromis_polyacanthus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSAMEG00000003119 | RAB11A | 84 | 100.000 | Ailuropoda_melanoleuca |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSACIG00000003092 | rab11a | 100 | 94.907 | Amphilophus_citrinellus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSAOCG00000020106 | rab11a | 100 | 94.907 | Amphiprion_ocellaris |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSAPEG00000000075 | rab11a | 100 | 94.907 | Amphiprion_percula |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSATEG00000022531 | rab11a | 100 | 94.907 | Anabas_testudineus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSAPLG00000014962 | RAB11A | 98 | 96.698 | Anas_platyrhynchos |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSACAG00000004479 | RAB11A | 99 | 99.070 | Anolis_carolinensis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSANAG00000008429 | RAB11A | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSACLG00000001775 | rab11a | 100 | 94.907 | Astatotilapia_calliptera |
| ENSG00000103769 | RAB11A | 95 | 100.000 | ENSAMXG00000003155 | rab11a | 97 | 93.780 | Astyanax_mexicanus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSBTAG00000018959 | RAB11A | 100 | 100.000 | Bos_taurus |
| ENSG00000103769 | RAB11A | 98 | 80.921 | ENSBTAG00000048711 | - | 97 | 79.310 | Bos_taurus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCJAG00000042037 | RAB11A | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCAFG00000017255 | RAB11A | 100 | 100.000 | Canis_familiaris |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCAFG00020008478 | RAB11A | 100 | 100.000 | Canis_lupus_dingo |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCHIG00000012183 | - | 100 | 100.000 | Capra_hircus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSTSYG00000031189 | RAB11A | 100 | 96.296 | Carlito_syrichta |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCPOG00000037599 | RAB11A | 100 | 100.000 | Cavia_porcellus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCCAG00000021286 | RAB11A | 100 | 100.000 | Cebus_capucinus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCATG00000034739 | - | 100 | 100.000 | Cercocebus_atys |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCLAG00000007435 | RAB11A | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCSAG00000013038 | RAB11A | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCPBG00000014614 | RAB11A | 100 | 99.537 | Chrysemys_picta_bellii |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCANG00000034570 | - | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCGRG00001023313 | Rab11a | 100 | 100.000 | Cricetulus_griseus_chok1gshd |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCGRG00000018359 | Rab11a | 100 | 100.000 | Cricetulus_griseus_crigri |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCSEG00000007383 | rab11a | 100 | 94.444 | Cynoglossus_semilaevis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSCVAG00000013878 | rab11a | 100 | 94.907 | Cyprinodon_variegatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSDARG00000041450 | rab11a | 100 | 95.370 | Danio_rerio |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSDNOG00000038479 | RAB11A | 83 | 100.000 | Dasypus_novemcinctus |
| ENSG00000103769 | RAB11A | 99 | 92.593 | FBgn0015790 | Rab11 | 100 | 85.514 | Drosophila_melanogaster |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSETEG00000004920 | RAB11A | 99 | 93.981 | Echinops_telfairi |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSEBUG00000007493 | rab11a | 100 | 88.018 | Eptatretus_burgeri |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSEASG00005006431 | RAB11A | 100 | 100.000 | Equus_asinus_asinus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSECAG00000023865 | RAB11A | 88 | 98.500 | Equus_caballus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSELUG00000002999 | rab11a | 100 | 95.370 | Esox_lucius |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSFCAG00000030174 | RAB11A | 100 | 100.000 | Felis_catus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSFALG00000010052 | RAB11A | 100 | 97.129 | Ficedula_albicollis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSFDAG00000021495 | RAB11A | 96 | 100.000 | Fukomys_damarensis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSFHEG00000019668 | rab11a | 100 | 94.907 | Fundulus_heteroclitus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGMOG00000004096 | - | 100 | 92.760 | Gadus_morhua |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGMOG00000008355 | rab11a | 99 | 93.056 | Gadus_morhua |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGALG00000007615 | RAB11A | 100 | 99.074 | Gallus_gallus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGAFG00000016549 | rab11a | 100 | 94.907 | Gambusia_affinis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGAGG00000014814 | RAB11A | 100 | 99.537 | Gopherus_agassizii |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSGGOG00000014252 | RAB11A | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000103769 | RAB11A | 100 | 91.358 | ENSHBUG00000023035 | rab11a | 88 | 94.089 | Haplochromis_burtoni |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSHGLG00000005396 | RAB11A | 100 | 100.000 | Heterocephalus_glaber_female |
| ENSG00000103769 | RAB11A | 100 | 87.654 | ENSHGLG00000006346 | - | 99 | 86.700 | Heterocephalus_glaber_female |
| ENSG00000103769 | RAB11A | 100 | 87.654 | ENSHGLG00100015502 | - | 99 | 86.700 | Heterocephalus_glaber_male |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSHGLG00100011237 | RAB11A | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSHCOG00000005652 | rab11a | 100 | 94.907 | Hippocampus_comes |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSIPUG00000021401 | Rab11a | 100 | 95.370 | Ictalurus_punctatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSTOG00000015255 | RAB11A | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSJJAG00000012036 | - | 100 | 100.000 | Jaculus_jaculus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSJJAG00000015445 | Rab11a | 100 | 100.000 | Jaculus_jaculus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSKMAG00000011905 | rab11a | 100 | 94.907 | Kryptolebias_marmoratus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSLBEG00000026857 | rab11a | 100 | 94.907 | Labrus_bergylta |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSLACG00000012184 | RAB11A | 100 | 95.833 | Latimeria_chalumnae |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSLOCG00000013835 | rab11a | 99 | 96.759 | Lepisosteus_oculatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSLAFG00000011418 | RAB11A | 100 | 100.000 | Loxodonta_africana |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMFAG00000000699 | - | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMMUG00000008006 | RAB11A | 100 | 100.000 | Macaca_mulatta |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMNEG00000043040 | - | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMLEG00000032505 | RAB11A | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMAMG00000017327 | rab11a | 100 | 94.907 | Mastacembelus_armatus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMZEG00005017063 | rab11a | 100 | 94.907 | Maylandia_zebra |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMGAG00000009797 | RAB11A | 100 | 97.585 | Meleagris_gallopavo |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMAUG00000018551 | Rab11a | 100 | 100.000 | Mesocricetus_auratus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMICG00000041504 | RAB11A | 100 | 100.000 | Microcebus_murinus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMOCG00000021138 | Rab11a | 100 | 100.000 | Microtus_ochrogaster |
| ENSG00000103769 | RAB11A | 99 | 95.425 | ENSMMOG00000017423 | - | 93 | 99.174 | Mola_mola |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMODG00000009810 | RAB11A | 100 | 100.000 | Monodelphis_domestica |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMALG00000003050 | rab11a | 100 | 94.907 | Monopterus_albus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | MGP_CAROLIEiJ_G0032346 | Rab11a | 100 | 100.000 | Mus_caroli |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMUSG00000004771 | Rab11a | 100 | 100.000 | Mus_musculus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | MGP_PahariEiJ_G0015211 | Rab11a | 100 | 100.000 | Mus_pahari |
| ENSG00000103769 | RAB11A | 100 | 100.000 | MGP_SPRETEiJ_G0033485 | Rab11a | 100 | 100.000 | Mus_spretus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMPUG00000000387 | RAB11A | 100 | 100.000 | Mustela_putorius_furo |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSMLUG00000008820 | RAB11A | 83 | 100.000 | Myotis_lucifugus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSNGAG00000013726 | Rab11a | 100 | 100.000 | Nannospalax_galili |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSNBRG00000012893 | rab11a | 100 | 94.907 | Neolamprologus_brichardi |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSNLEG00000012853 | - | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOPRG00000004681 | RAB11A | 100 | 97.222 | Ochotona_princeps |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSODEG00000016878 | - | 100 | 100.000 | Octodon_degus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSONIG00000015072 | RAB11A | 100 | 94.907 | Oreochromis_niloticus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOANG00000002201 | RAB11A | 90 | 100.000 | Ornithorhynchus_anatinus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOCUG00000014755 | RAB11A | 100 | 100.000 | Oryctolagus_cuniculus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSORLG00000018331 | RAB11A | 100 | 95.370 | Oryzias_latipes |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSORLG00015010257 | rab11a | 100 | 95.370 | Oryzias_latipes_hsok |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOMEG00000017467 | rab11a | 100 | 95.370 | Oryzias_melastigma |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOGAG00000007642 | RAB11A | 100 | 100.000 | Otolemur_garnettii |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSOARG00000018084 | RAB11A | 83 | 100.000 | Ovis_aries |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPPAG00000030195 | - | 100 | 100.000 | Pan_paniscus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPPRG00000010471 | RAB11A | 100 | 100.000 | Panthera_pardus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPTIG00000021637 | RAB11A | 100 | 100.000 | Panthera_tigris_altaica |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPTRG00000044358 | - | 100 | 100.000 | Pan_troglodytes |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPANG00000016687 | - | 100 | 100.000 | Papio_anubis |
| ENSG00000103769 | RAB11A | 100 | 96.296 | ENSPKIG00000015102 | - | 100 | 96.296 | Paramormyrops_kingsleyae |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPKIG00000007606 | rab11a | 100 | 94.907 | Paramormyrops_kingsleyae |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPSIG00000011930 | RAB11A | 97 | 97.143 | Pelodiscus_sinensis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPEMG00000021125 | - | 100 | 100.000 | Peromyscus_maniculatus_bairdii |
| ENSG00000103769 | RAB11A | 100 | 98.765 | ENSPEMG00000005147 | - | 98 | 99.510 | Peromyscus_maniculatus_bairdii |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPCIG00000006878 | RAB11A | 93 | 100.000 | Phascolarctos_cinereus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPFOG00000009284 | rab11a | 100 | 94.907 | Poecilia_formosa |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPLAG00000011175 | rab11a | 100 | 94.907 | Poecilia_latipinna |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPMEG00000007932 | rab11a | 100 | 94.907 | Poecilia_mexicana |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPREG00000009605 | rab11a | 100 | 94.907 | Poecilia_reticulata |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPPYG00000006573 | RAB11A | 100 | 100.000 | Pongo_abelii |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPCAG00000002640 | RAB11A | 100 | 99.537 | Procavia_capensis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPCOG00000015802 | - | 100 | 99.537 | Propithecus_coquereli |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPCOG00000016536 | - | 100 | 95.833 | Propithecus_coquereli |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPNYG00000002252 | rab11a | 100 | 94.907 | Pundamilia_nyererei |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSPNAG00000014863 | rab11a | 100 | 95.370 | Pygocentrus_nattereri |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSRNOG00000011302 | Rab11a | 100 | 100.000 | Rattus_norvegicus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSRBIG00000035649 | RAB11A | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSRROG00000035305 | RAB11A | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSBOG00000017326 | RAB11A | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSHAG00000006622 | RAB11A | 97 | 97.129 | Sarcophilus_harrisii |
| ENSG00000103769 | RAB11A | 100 | 93.981 | ENSSFOG00015024042 | rab11a | 100 | 93.981 | Scleropages_formosus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSMAG00000004133 | rab11a | 100 | 94.907 | Scophthalmus_maximus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSDUG00000002580 | rab11a | 100 | 94.907 | Seriola_dumerili |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSLDG00000009438 | rab11a | 100 | 94.907 | Seriola_lalandi_dorsalis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSPUG00000013698 | RAB11A | 100 | 99.507 | Sphenodon_punctatus |
| ENSG00000103769 | RAB11A | 99 | 95.425 | ENSSPAG00000019393 | RAB11A | 87 | 95.455 | Stegastes_partitus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSSSCG00000032312 | RAB11A | 100 | 100.000 | Sus_scrofa |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSTGUG00000009279 | RAB11A | 100 | 99.074 | Taeniopygia_guttata |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSTRUG00000020792 | rab11a | 100 | 94.907 | Takifugu_rubripes |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSTTRG00000011885 | RAB11A | 100 | 98.140 | Tursiops_truncatus |
| ENSG00000103769 | RAB11A | 100 | 98.765 | ENSUAMG00000026548 | RAB11A | 98 | 98.544 | Ursus_americanus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSUMAG00000012273 | RAB11A | 100 | 100.000 | Ursus_maritimus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSVPAG00000004556 | RAB11A | 100 | 100.000 | Vicugna_pacos |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSVVUG00000014435 | RAB11A | 100 | 100.000 | Vulpes_vulpes |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSXETG00000006639 | rab11a | 100 | 98.148 | Xenopus_tropicalis |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSXCOG00000001357 | rab11a | 82 | 94.907 | Xiphophorus_couchianus |
| ENSG00000103769 | RAB11A | 100 | 100.000 | ENSXMAG00000023732 | rab11a | 82 | 94.907 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000922 | spindle pole | 24561039. | IDA | Component |
| GO:0003091 | renal water homeostasis | - | TAS | Process |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0003924 | GTPase activity | 15837192.16905101.17007872.17030804.17562788. | IDA | Function |
| GO:0005515 | protein binding | 12470645.15601896.16148947.16730941.16905101.17007872.17030804.17082457.17229837.17462998.17562788.18329369.18511905.19542231.20308558.21976701.23535298.24591568.24876499.25556234.26258637.26496610.28468127.29568061.29997244.30217979. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005768 | endosome | 21873635. | IBA | Component |
| GO:0005771 | multivesicular body | 15634213. | IDA | Component |
| GO:0005794 | Golgi apparatus | 16791210. | HDA | Component |
| GO:0005802 | trans-Golgi network | 15229288. | IDA | Component |
| GO:0005813 | centrosome | 24648492. | IDA | Component |
| GO:0005815 | microtubule organizing center | - | IDA | Component |
| GO:0005828 | kinetochore microtubule | 24561039. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0006887 | exocytosis | 21873635. | IBA | Process |
| GO:0007080 | mitotic metaphase plate congression | 24561039. | IMP | Process |
| GO:0008017 | microtubule binding | 24561039. | IDA | Function |
| GO:0010634 | positive regulation of epithelial cell migration | 22613965. | IMP | Process |
| GO:0010796 | regulation of multivesicular body size | 15634213. | IMP | Process |
| GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 24561039. | IMP | Process |
| GO:0016192 | vesicle-mediated transport | 21873635. | IBA | Process |
| GO:0016192 | vesicle-mediated transport | 17462998.19542231. | IDA | Process |
| GO:0019905 | syntaxin binding | 12145319. | NAS | Function |
| GO:0030133 | transport vesicle | 21873635. | IBA | Component |
| GO:0030424 | axon | 21873635. | IBA | Component |
| GO:0030659 | cytoplasmic vesicle membrane | - | TAS | Component |
| GO:0030953 | astral microtubule organization | 24561039. | IMP | Process |
| GO:0031175 | neuron projection development | 17082457. | IMP | Process |
| GO:0031410 | cytoplasmic vesicle | 22613965. | IDA | Component |
| GO:0031489 | myosin V binding | 24006491. | IPI | Function |
| GO:0031982 | vesicle | 18570918. | IDA | Component |
| GO:0032154 | cleavage furrow | 15601896. | IDA | Component |
| GO:0032402 | melanosome transport | 21873635. | IBA | Process |
| GO:0032402 | melanosome transport | - | ISS | Process |
| GO:0032465 | regulation of cytokinesis | 15601896. | IMP | Process |
| GO:0032482 | Rab protein signal transduction | 21873635. | IBA | Process |
| GO:0032991 | protein-containing complex | 17462998. | IDA | Component |
| GO:0036258 | multivesicular body assembly | 15634213. | IMP | Process |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0043687 | post-translational protein modification | - | TAS | Process |
| GO:0045335 | phagocytic vesicle | 21255211. | IDA | Component |
| GO:0045773 | positive regulation of axon extension | - | IEA | Process |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | - | IEA | Process |
| GO:0048227 | plasma membrane to endosome transport | 11163216. | NAS | Process |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0051223 | regulation of protein transport | - | IEA | Process |
| GO:0051650 | establishment of vesicle localization | 24561039. | IMP | Process |
| GO:0055037 | recycling endosome | 21873635. | IBA | Component |
| GO:0055037 | recycling endosome | 24561039. | IDA | Component |
| GO:0055037 | recycling endosome | - | ISS | Component |
| GO:0055038 | recycling endosome membrane | - | TAS | Component |
| GO:0060627 | regulation of vesicle-mediated transport | 22613965. | IMP | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:0070062 | extracellular exosome | 15326289. | IDA | Component |
| GO:0072594 | establishment of protein localization to organelle | 24561039. | IMP | Process |
| GO:0072659 | protein localization to plasma membrane | 17082457. | IDA | Process |
| GO:0090150 | establishment of protein localization to membrane | 24006491. | IMP | Process |
| GO:0090307 | mitotic spindle assembly | 24561039. | IMP | Process |
| GO:0097711 | ciliary basal body-plasma membrane docking | - | TAS | Process |
| GO:0098685 | Schaffer collateral - CA1 synapse | - | IEA | Component |
| GO:0098837 | postsynaptic recycling endosome | 26565907. | EXP | Component |
| GO:0098837 | postsynaptic recycling endosome | 21873635. | IBA | Component |
| GO:0098837 | postsynaptic recycling endosome | 26565907. | IDA | Component |
| GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 21873635. | IBA | Process |
| GO:0098978 | glutamatergic synapse | 26565907. | EXP | Component |
| GO:0098978 | glutamatergic synapse | 26565907. | IDA | Component |
| GO:0099003 | vesicle-mediated transport in synapse | - | IEA | Process |
| GO:0150093 | amyloid-beta clearance by transcytosis | 26005850. | IGI | Process |
| GO:1903078 | positive regulation of protein localization to plasma membrane | - | IEA | Process |
| GO:1990182 | exosomal secretion | 15634213. | IMP | Process |