
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000600909 | KCNN4-207 | 1190 | - | ENSP00000470339 | 59 (aa) | - | M0QZ70 |
| ENST00000615047 | KCNN4-209 | 2029 | XM_005258883 | ENSP00000485014 | 295 (aa) | XP_005258940 | D1MQ08 |
| ENST00000601549 | KCNN4-208 | 1406 | - | - | - (aa) | - | - |
| ENST00000262888 | KCNN4-201 | 2244 | XM_005258882 | ENSP00000262888 | 427 (aa) | XP_005258939 | O15554 |
| ENST00000598836 | KCNN4-203 | 1003 | - | ENSP00000471900 | 165 (aa) | - | M0R1J0 |
| ENST00000648053 | KCNN4-210 | 1270 | - | - | - (aa) | - | - |
| ENST00000599720 | KCNN4-205 | 983 | - | ENSP00000472513 | 57 (aa) | - | M0R2F1 |
| ENST00000600408 | KCNN4-206 | 648 | - | ENSP00000472510 | 113 (aa) | - | M0R2E8 |
| ENST00000648319 | KCNN4-211 | 1956 | - | ENSP00000496939 | 427 (aa) | - | UPI0000000DF3 |
| ENST00000597184 | KCNN4-202 | 631 | - | - | - (aa) | - | - |
| ENST00000599107 | KCNN4-204 | 687 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000104783 | Breast Neoplasms | 2E-10 | 23535729 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000104783 | rs56344893 | 19 | 43778377 | A | Monocyte count | 27863252 | [0.04-0.055] unit decrease | 0.04736383 | EFO_0005091 |
| ENSG00000104783 | rs563995 | 19 | 43774629 | A | Mean corpuscular volume | 27863252 | [0.017-0.034] unit decrease | 0.02572105 | EFO_0004526 |
| ENSG00000104783 | rs11879798 | 19 | 43777746 | A | Granulocyte percentage of myeloid white cells | 27863252 | [0.038-0.053] unit increase | 0.04540951 | EFO_0007997 |
| ENSG00000104783 | rs563995 | 19 | 43774629 | A | Hemoglobin concentration | 27863252 | [0.017-0.034] unit increase | 0.02525654 | EFO_0004509 |
| ENSG00000104783 | rs11879798 | 19 | 43777746 | A | Monocyte percentage of white cells | 27863252 | [0.04-0.054] unit decrease | 0.04723772 | EFO_0007989 |
| ENSG00000104783 | rs563995 | 19 | 43774629 | A | Mean corpuscular hemoglobin concentration | 27863252 | [0.037-0.054] unit increase | 0.04561461 | EFO_0004528 |
| ENSG00000104783 | rs649540 | 19 | 43774627 | ? | White blood cell count | 30595370 | EFO_0004308 | ||
| ENSG00000104783 | rs347519 | 19 | 43769364 | ? | Red blood cell count | 30595370 | EFO_0004305 | ||
| ENSG00000104783 | rs1685191 | 19 | 43779080 | T | Breast cancer | 29059683 | [0.046-0.071] unit decrease | 0.0586 | EFO_0000305 |
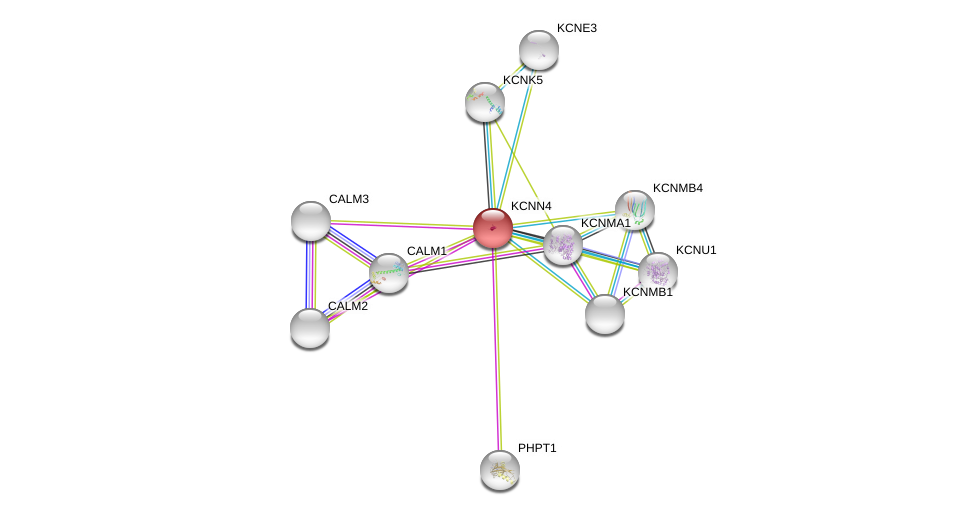
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002376 | immune system process | - | IEA | Process |
| GO:0005267 | potassium channel activity | - | TAS | Function |
| GO:0005515 | protein binding | 10880439.12198491.18796614.27092946.29724949. | IPI | Function |
| GO:0005516 | calmodulin binding | 21873635. | IBA | Function |
| GO:0005886 | plasma membrane | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | 26148990. | IDA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0006813 | potassium ion transport | 9380751. | TAS | Process |
| GO:0006816 | calcium ion transport | 18796614. | IDA | Process |
| GO:0006884 | cell volume homeostasis | - | IEA | Process |
| GO:0006952 | defense response | 9407042. | TAS | Process |
| GO:0008076 | voltage-gated potassium channel complex | 18796614. | IC | Component |
| GO:0015269 | calcium-activated potassium channel activity | 18796614.26148990. | IDA | Function |
| GO:0016286 | small conductance calcium-activated potassium channel activity | - | IEA | Function |
| GO:0019903 | protein phosphatase binding | 18796614. | IPI | Function |
| GO:0022894 | Intermediate conductance calcium-activated potassium channel activity | 21873635. | IBA | Function |
| GO:0030322 | stabilization of membrane potential | 18796614. | IDA | Process |
| GO:0031982 | vesicle | 25468996. | IDA | Component |
| GO:0043005 | neuron projection | 21873635. | IBA | Component |
| GO:0043025 | neuronal cell body | 21873635. | IBA | Component |
| GO:0045332 | phospholipid translocation | - | IEA | Process |
| GO:0046541 | saliva secretion | - | IEA | Process |
| GO:0050714 | positive regulation of protein secretion | - | IEA | Process |
| GO:0050862 | positive regulation of T cell receptor signaling pathway | 18796614. | IDA | Process |
| GO:0071805 | potassium ion transmembrane transport | 21873635. | IBA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| READ | |||
| SKCM | |||
| TGCT | |||
| THCA | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| MESO | ||
| SKCM | ||
| PRAD | ||
| LUSC | ||
| BRCA | ||
| TGCT | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| UCEC | ||
| GBM | ||
| DLBC | ||
| LUAD |