
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000221448 | RRM_1 | PF00076.22 | 2.1e-17 | 1 | 1 |
| ENSP00000472998 | RRM_1 | PF00076.22 | 2.2e-17 | 1 | 1 |
| ENSP00000470023 | RRM_1 | PF00076.22 | 2.4e-13 | 1 | 1 |
| ENSP00000221448 | U1snRNP70_N | PF12220.8 | 3.8e-27 | 1 | 1 |
| ENSP00000472998 | U1snRNP70_N | PF12220.8 | 3.9e-27 | 1 | 1 |
| ENSP00000468828 | U1snRNP70_N | PF12220.8 | 0.00027 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000598984 | SNRNP70-207 | 547 | - | ENSP00000470023 | 94 (aa) | - | M0QYR1 |
| ENST00000601411 | SNRNP70-210 | 377 | - | ENSP00000468828 | 33 (aa) | - | M0QX04 |
| ENST00000544278 | SNRNP70-203 | 3221 | - | - | - (aa) | - | - |
| ENST00000595231 | SNRNP70-204 | 1673 | - | ENSP00000471006 | 166 (aa) | - | P08621 |
| ENST00000401730 | SNRNP70-202 | 3144 | - | ENSP00000385077 | 166 (aa) | - | P08621 |
| ENST00000221448 | SNRNP70-201 | 1603 | - | ENSP00000221448 | 428 (aa) | - | P08621 |
| ENST00000597936 | SNRNP70-205 | 580 | - | - | - (aa) | - | - |
| ENST00000599687 | SNRNP70-208 | 563 | - | - | - (aa) | - | - |
| ENST00000598441 | SNRNP70-206 | 1574 | - | ENSP00000472998 | 437 (aa) | - | P08621 |
| ENST00000601065 | SNRNP70-209 | 1757 | - | ENSP00000468952 | 166 (aa) | - | P08621 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03040 | Spliceosome | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000104852 | rs73046792 | 19 | 49102448 | ? | Systolic blood pressure | 30595370 | EFO_0006335 | ||
| ENSG00000104852 | rs73046792 | 19 | 49102448 | G | Systolic blood pressure | 30224653 | [0.27-0.44] mmHg increase | 0.3554 | EFO_0006335 |
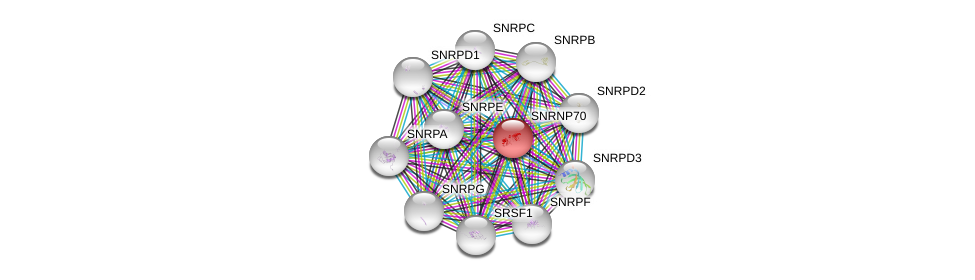
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000104852 | SNRNP70 | 55 | 36.538 | ENSG00000242389 | RBMY1E | 85 | 34.286 |
| ENSG00000104852 | SNRNP70 | 60 | 39.286 | ENSG00000188529 | SRSF10 | 81 | 39.286 |
| ENSG00000104852 | SNRNP70 | 55 | 36.538 | ENSG00000234414 | RBMY1A1 | 85 | 34.286 |
| ENSG00000104852 | SNRNP70 | 55 | 36.538 | ENSG00000244395 | RBMY1D | 85 | 34.286 |
| ENSG00000104852 | SNRNP70 | 55 | 36.538 | ENSG00000242875 | RBMY1B | 85 | 34.286 |
| ENSG00000104852 | SNRNP70 | 51 | 37.500 | ENSG00000132819 | RBM38 | 63 | 32.468 |
| ENSG00000104852 | SNRNP70 | 55 | 32.692 | ENSG00000099622 | CIRBP | 91 | 34.247 |
| ENSG00000104852 | SNRNP70 | 51 | 41.667 | ENSG00000265241 | RBM8A | 70 | 33.784 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000104852 | SNRNP70 | 86 | 88.028 | ENSAPOG00000009322 | snrnp70 | 89 | 88.028 | Acanthochromis_polyacanthus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSAMEG00000014468 | SNRNP70 | 100 | 93.878 | Ailuropoda_melanoleuca |
| ENSG00000104852 | SNRNP70 | 99 | 89.634 | ENSAOCG00000011672 | snrnp70 | 50 | 83.482 | Amphiprion_ocellaris |
| ENSG00000104852 | SNRNP70 | 99 | 93.902 | ENSACAG00000013276 | SNRNP70 | 51 | 97.297 | Anolis_carolinensis |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSANAG00000036787 | SNRNP70 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSBTAG00000048800 | - | 100 | 100.000 | Bos_taurus |
| ENSG00000104852 | SNRNP70 | 99 | 52.688 | WBGene00004390 | rnp-7 | 56 | 53.723 | Caenorhabditis_elegans |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCJAG00000000193 | SNRNP70 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCAFG00000003711 | SNRNP70 | 100 | 100.000 | Canis_familiaris |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCAFG00020025470 | SNRNP70 | 100 | 96.388 | Canis_lupus_dingo |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCHIG00000018534 | - | 100 | 100.000 | Capra_hircus |
| ENSG00000104852 | SNRNP70 | 63 | 88.571 | ENSTSYG00000032350 | - | 85 | 88.571 | Carlito_syrichta |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCPOG00000019589 | SNRNP70 | 100 | 97.059 | Cavia_porcellus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCCAG00000036551 | SNRNP70 | 100 | 99.486 | Cebus_capucinus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCATG00000016149 | SNRNP70 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCLAG00000007991 | SNRNP70 | 100 | 98.178 | Chinchilla_lanigera |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCSAG00000002759 | SNRNP70 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000104852 | SNRNP70 | 100 | 96.970 | ENSCPBG00000014841 | SNRNP70 | 51 | 99.099 | Chrysemys_picta_bellii |
| ENSG00000104852 | SNRNP70 | 97 | 71.429 | ENSCSAVG00000010109 | - | 50 | 70.681 | Ciona_savignyi |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCANG00000036607 | SNRNP70 | 100 | 99.772 | Colobus_angolensis_palliatus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCGRG00001020676 | Snrnp70 | 100 | 95.973 | Cricetulus_griseus_chok1gshd |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSCGRG00000018048 | Snrnp70 | 100 | 95.973 | Cricetulus_griseus_crigri |
| ENSG00000104852 | SNRNP70 | 99 | 88.415 | ENSCSEG00000011978 | snrnp70 | 99 | 91.480 | Cynoglossus_semilaevis |
| ENSG00000104852 | SNRNP70 | 96 | 89.937 | ENSCVAG00000003912 | snrnp70 | 99 | 89.937 | Cyprinodon_variegatus |
| ENSG00000104852 | SNRNP70 | 99 | 86.585 | ENSDARG00000077126 | snrnp70 | 99 | 88.820 | Danio_rerio |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSDNOG00000035849 | - | 100 | 100.000 | Dasypus_novemcinctus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSDORG00000010228 | Snrnp70 | 100 | 95.946 | Dipodomys_ordii |
| ENSG00000104852 | SNRNP70 | 74 | 100.000 | ENSETEG00000005009 | SNRNP70 | 74 | 100.000 | Echinops_telfairi |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSEASG00005004917 | SNRNP70 | 100 | 97.955 | Equus_asinus_asinus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSECAG00000013936 | - | 100 | 100.000 | Equus_caballus |
| ENSG00000104852 | SNRNP70 | 100 | 97.872 | ENSEEUG00000005666 | SNRNP70 | 100 | 82.232 | Erinaceus_europaeus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSFCAG00000042603 | SNRNP70 | 100 | 91.591 | Felis_catus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSFDAG00000013830 | SNRNP70 | 96 | 99.682 | Fukomys_damarensis |
| ENSG00000104852 | SNRNP70 | 99 | 87.805 | ENSGMOG00000011492 | snrnp70 | 97 | 91.403 | Gadus_morhua |
| ENSG00000104852 | SNRNP70 | 99 | 88.415 | ENSGACG00000000448 | snrnp70 | 70 | 91.928 | Gasterosteus_aculeatus |
| ENSG00000104852 | SNRNP70 | 96 | 95.556 | ENSGAGG00000012628 | SNRNP70 | 99 | 80.000 | Gopherus_agassizii |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSGGOG00000008245 | SNRNP70 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSHGLG00100001651 | - | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSSTOG00000013988 | - | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSJJAG00000002614 | Snrnp70 | 100 | 95.291 | Jaculus_jaculus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSLAFG00000028536 | SNRNP70 | 100 | 100.000 | Loxodonta_africana |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMFAG00000039824 | SNRNP70 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMMUG00000004928 | SNRNP70 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMNEG00000038057 | SNRNP70 | 100 | 98.974 | Macaca_nemestrina |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMLEG00000030967 | SNRNP70 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMAUG00000020970 | Snrnp70 | 100 | 96.188 | Mesocricetus_auratus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMICG00000038437 | SNRNP70 | 100 | 100.000 | Microcebus_murinus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMOCG00000012825 | Snrnp70 | 100 | 95.323 | Microtus_ochrogaster |
| ENSG00000104852 | SNRNP70 | 75 | 76.716 | ENSMODG00000023381 | SNRNP70 | 100 | 80.000 | Monodelphis_domestica |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | MGP_CAROLIEiJ_G0029714 | Snrnp70 | 100 | 100.000 | Mus_caroli |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMUSG00000063511 | Snrnp70 | 100 | 100.000 | Mus_musculus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | MGP_PahariEiJ_G0012939 | Snrnp70 | 100 | 100.000 | Mus_pahari |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | MGP_SPRETEiJ_G0030812 | Snrnp70 | 100 | 100.000 | Mus_spretus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMPUG00000003258 | SNRNP70 | 100 | 98.182 | Mustela_putorius_furo |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSMLUG00000011766 | SNRNP70 | 100 | 98.182 | Myotis_lucifugus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSNGAG00000021191 | Snrnp70 | 95 | 97.531 | Nannospalax_galili |
| ENSG00000104852 | SNRNP70 | 99 | 89.024 | ENSNBRG00000003126 | snrnp70 | 96 | 92.377 | Neolamprologus_brichardi |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSNLEG00000004899 | SNRNP70 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSODEG00000017499 | SNRNP70 | 100 | 93.290 | Octodon_degus |
| ENSG00000104852 | SNRNP70 | 99 | 88.415 | ENSONIG00000007822 | snrnp70 | 52 | 91.928 | Oreochromis_niloticus |
| ENSG00000104852 | SNRNP70 | 100 | 97.872 | ENSOANG00000005213 | SNRNP70 | 97 | 99.550 | Ornithorhynchus_anatinus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSOCUG00000015081 | SNRNP70 | 85 | 92.268 | Oryctolagus_cuniculus |
| ENSG00000104852 | SNRNP70 | 100 | 97.895 | ENSOGAG00000029310 | SNRNP70 | 100 | 97.511 | Otolemur_garnettii |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSOARG00000012608 | SNRNP70 | 93 | 100.000 | Ovis_aries |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPPAG00000035067 | SNRNP70 | 100 | 100.000 | Pan_paniscus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPPRG00000005579 | SNRNP70 | 100 | 98.182 | Panthera_pardus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPTRG00000011277 | SNRNP70 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPANG00000004068 | SNRNP70 | 100 | 100.000 | Papio_anubis |
| ENSG00000104852 | SNRNP70 | 96 | 80.503 | ENSPKIG00000019992 | snrnp70 | 86 | 79.012 | Paramormyrops_kingsleyae |
| ENSG00000104852 | SNRNP70 | 100 | 96.970 | ENSPSIG00000015251 | SNRNP70 | 90 | 85.217 | Pelodiscus_sinensis |
| ENSG00000104852 | SNRNP70 | 96 | 90.566 | ENSPMGG00000010153 | snrnp70 | 77 | 90.062 | Periophthalmus_magnuspinnatus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPEMG00000019011 | Snrnp70 | 100 | 95.333 | Peromyscus_maniculatus_bairdii |
| ENSG00000104852 | SNRNP70 | 99 | 79.878 | ENSPMAG00000008502 | snrnp70 | 97 | 83.422 | Petromyzon_marinus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPCIG00000007296 | SNRNP70 | 100 | 88.018 | Phascolarctos_cinereus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSPPYG00000010234 | SNRNP70 | 100 | 99.542 | Pongo_abelii |
| ENSG00000104852 | SNRNP70 | 99 | 85.976 | ENSPCOG00000023103 | SNRNP70 | 100 | 90.787 | Propithecus_coquereli |
| ENSG00000104852 | SNRNP70 | 100 | 94.545 | ENSPVAG00000009578 | SNRNP70 | 100 | 92.955 | Pteropus_vampyrus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSRBIG00000042387 | SNRNP70 | 100 | 99.771 | Rhinopithecus_bieti |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSRROG00000033636 | SNRNP70 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSSBOG00000020026 | SNRNP70 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSSHAG00000002194 | SNRNP70 | 100 | 88.710 | Sarcophilus_harrisii |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSSPUG00000016621 | SNRNP70 | 52 | 98.649 | Sphenodon_punctatus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSSSCG00000023792 | - | 100 | 100.000 | Sus_scrofa |
| ENSG00000104852 | SNRNP70 | 52 | 93.878 | ENSTGUG00000016869 | - | 100 | 92.157 | Taeniopygia_guttata |
| ENSG00000104852 | SNRNP70 | 66 | 90.000 | ENSTNIG00000014186 | - | 99 | 90.000 | Tetraodon_nigroviridis |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSTTRG00000017099 | SNRNP70 | 100 | 97.506 | Tursiops_truncatus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSUAMG00000016147 | SNRNP70 | 100 | 98.182 | Ursus_americanus |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSUMAG00000001467 | SNRNP70 | 100 | 100.000 | Ursus_maritimus |
| ENSG00000104852 | SNRNP70 | 100 | 96.970 | ENSVPAG00000005959 | - | 100 | 80.995 | Vicugna_pacos |
| ENSG00000104852 | SNRNP70 | 100 | 100.000 | ENSVVUG00000022976 | SNRNP70 | 100 | 100.000 | Vulpes_vulpes |
| ENSG00000104852 | SNRNP70 | 99 | 93.293 | ENSXETG00000022919 | snrnp70 | 57 | 95.595 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | 21873635. | IBA | Process |
| GO:0000398 | mRNA splicing, via spliceosome | 9731529. | IC | Process |
| GO:0000398 | mRNA splicing, via spliceosome | 9531537. | IDA | Process |
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 2467746. | IMP | Function |
| GO:0003729 | mRNA binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 9447963.14561889.18559666.18951082.19928837.21988832.22365833.23602568.23793891.25719862.26124092.26828962. | IPI | Function |
| GO:0005634 | nucleus | 18559850.21113136. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005681 | spliceosomal complex | 9531537.9731529. | IDA | Component |
| GO:0005685 | U1 snRNP | 21873635. | IBA | Component |
| GO:0005685 | U1 snRNP | 21113136.23793891.25555158. | IDA | Component |
| GO:0016607 | nuclear speck | - | ISS | Component |
| GO:0030619 | U1 snRNA binding | 21113136.25555158. | IDA | Function |
| GO:0043462 | regulation of ATPase activity | 25719862. | TAS | Process |
| GO:0043484 | regulation of RNA splicing | 2467746. | IMP | Process |
| GO:0048026 | positive regulation of mRNA splicing, via spliceosome | - | IEA | Process |
| GO:0061084 | negative regulation of protein refolding | 25719862. | TAS | Process |
| GO:1904715 | negative regulation of chaperone-mediated autophagy | 25719862. | TAS | Process |