
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000269701 | AKAP95 | PF04988.12 | 5.9e-67 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 26683827 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000599883 | AKAP8-205 | 860 | - | ENSP00000468902 | 133 (aa) | - | M0QX51 |
| ENST00000537303 | AKAP8-202 | 1350 | - | - | - (aa) | - | - |
| ENST00000595416 | AKAP8-203 | 557 | - | - | - (aa) | - | - |
| ENST00000598597 | AKAP8-204 | 3071 | - | ENSP00000469908 | 178 (aa) | - | M0QYL4 |
| ENST00000269701 | AKAP8-201 | 3670 | XM_011527624 | ENSP00000269701 | 692 (aa) | XP_011525926 | O43823 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000105127 | rs144075571 | 19 | 15379590 | G | RANTES levels | 27989323 | [0.22-0.55] SD units increase | 0.3831 | EFO_0005117 |
| ENSG00000105127 | rs143155169 | 19 | 15367524 | ? | Red blood cell count | 30595370 | EFO_0004305 |
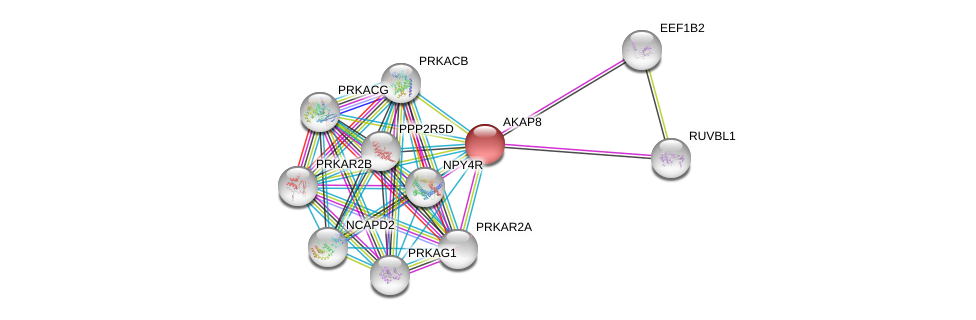
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000105127 | AKAP8 | 66 | 37.097 | ENSG00000011243 | AKAP8L | 91 | 33.509 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000105127 | AKAP8 | 100 | 83.429 | ENSAMEG00000002281 | AKAP8 | 100 | 82.853 | Ailuropoda_melanoleuca |
| ENSG00000105127 | AKAP8 | 66 | 35.211 | ENSAMEG00000002248 | AKAP8L | 84 | 32.821 | Ailuropoda_melanoleuca |
| ENSG00000105127 | AKAP8 | 53 | 35.450 | ENSAOCG00000022319 | akap8l | 86 | 30.349 | Amphiprion_ocellaris |
| ENSG00000105127 | AKAP8 | 53 | 35.450 | ENSAPEG00000009544 | akap8l | 86 | 30.349 | Amphiprion_percula |
| ENSG00000105127 | AKAP8 | 51 | 36.141 | ENSATEG00000003803 | akap8l | 93 | 30.737 | Anabas_testudineus |
| ENSG00000105127 | AKAP8 | 99 | 55.114 | ENSAPLG00000007188 | AKAP8 | 99 | 54.622 | Anas_platyrhynchos |
| ENSG00000105127 | AKAP8 | 62 | 41.593 | ENSAPLG00000013599 | AKAP8L | 79 | 39.456 | Anas_platyrhynchos |
| ENSG00000105127 | AKAP8 | 95 | 44.944 | ENSACAG00000017298 | AKAP8 | 100 | 54.849 | Anolis_carolinensis |
| ENSG00000105127 | AKAP8 | 62 | 37.849 | ENSACAG00000017315 | AKAP8L | 82 | 37.500 | Anolis_carolinensis |
| ENSG00000105127 | AKAP8 | 66 | 36.364 | ENSANAG00000036794 | AKAP8L | 95 | 33.095 | Aotus_nancymaae |
| ENSG00000105127 | AKAP8 | 100 | 90.607 | ENSANAG00000036569 | AKAP8 | 100 | 90.751 | Aotus_nancymaae |
| ENSG00000105127 | AKAP8 | 78 | 34.075 | ENSBTAG00000010439 | AKAP8L | 86 | 33.703 | Bos_taurus |
| ENSG00000105127 | AKAP8 | 66 | 37.349 | ENSCJAG00000002227 | AKAP8L | 95 | 33.429 | Callithrix_jacchus |
| ENSG00000105127 | AKAP8 | 100 | 89.017 | ENSCJAG00000002212 | AKAP8 | 100 | 89.162 | Callithrix_jacchus |
| ENSG00000105127 | AKAP8 | 100 | 81.339 | ENSCAFG00000016083 | AKAP8 | 100 | 81.197 | Canis_familiaris |
| ENSG00000105127 | AKAP8 | 66 | 35.743 | ENSCAFG00000016050 | AKAP8L | 80 | 33.853 | Canis_familiaris |
| ENSG00000105127 | AKAP8 | 99 | 77.364 | ENSCAFG00020009384 | AKAP8 | 99 | 77.077 | Canis_lupus_dingo |
| ENSG00000105127 | AKAP8 | 66 | 35.671 | ENSCAFG00020009405 | AKAP8L | 86 | 33.798 | Canis_lupus_dingo |
| ENSG00000105127 | AKAP8 | 71 | 34.831 | ENSCHIG00000005712 | AKAP8L | 81 | 33.795 | Capra_hircus |
| ENSG00000105127 | AKAP8 | 99 | 76.890 | ENSCHIG00000008591 | AKAP8 | 99 | 77.536 | Capra_hircus |
| ENSG00000105127 | AKAP8 | 100 | 86.455 | ENSTSYG00000009048 | AKAP8 | 100 | 86.888 | Carlito_syrichta |
| ENSG00000105127 | AKAP8 | 66 | 35.872 | ENSTSYG00000030638 | AKAP8L | 87 | 33.438 | Carlito_syrichta |
| ENSG00000105127 | AKAP8 | 61 | 32.222 | ENSCAPG00000011498 | AKAP8L | 81 | 31.332 | Cavia_aperea |
| ENSG00000105127 | AKAP8 | 85 | 85.932 | ENSCAPG00000008969 | AKAP8 | 99 | 78.696 | Cavia_aperea |
| ENSG00000105127 | AKAP8 | 75 | 33.102 | ENSCPOG00000036348 | AKAP8L | 80 | 33.746 | Cavia_porcellus |
| ENSG00000105127 | AKAP8 | 99 | 79.476 | ENSCPOG00000009712 | AKAP8 | 99 | 79.592 | Cavia_porcellus |
| ENSG00000105127 | AKAP8 | 66 | 36.895 | ENSCCAG00000036248 | AKAP8L | 91 | 33.934 | Cebus_capucinus |
| ENSG00000105127 | AKAP8 | 96 | 90.058 | ENSCCAG00000037134 | AKAP8 | 100 | 85.694 | Cebus_capucinus |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSCATG00000039160 | AKAP8L | 95 | 33.286 | Cercocebus_atys |
| ENSG00000105127 | AKAP8 | 96 | 95.322 | ENSCATG00000043874 | AKAP8 | 100 | 89.306 | Cercocebus_atys |
| ENSG00000105127 | AKAP8 | 100 | 83.718 | ENSCLAG00000008154 | AKAP8 | 100 | 84.438 | Chinchilla_lanigera |
| ENSG00000105127 | AKAP8 | 66 | 35.412 | ENSCLAG00000008083 | AKAP8L | 95 | 33.619 | Chinchilla_lanigera |
| ENSG00000105127 | AKAP8 | 65 | 36.864 | ENSCSAG00000006087 | AKAP8L | 99 | 33.233 | Chlorocebus_sabaeus |
| ENSG00000105127 | AKAP8 | 96 | 94.737 | ENSCSAG00000006104 | AKAP8 | 100 | 97.412 | Chlorocebus_sabaeus |
| ENSG00000105127 | AKAP8 | 69 | 39.010 | ENSCPBG00000020814 | AKAP8L | 80 | 38.417 | Chrysemys_picta_bellii |
| ENSG00000105127 | AKAP8 | 81 | 55.863 | ENSCPBG00000020818 | AKAP8 | 99 | 50.988 | Chrysemys_picta_bellii |
| ENSG00000105127 | AKAP8 | 66 | 35.542 | ENSCANG00000037601 | AKAP8L | 90 | 33.675 | Colobus_angolensis_palliatus |
| ENSG00000105127 | AKAP8 | 96 | 95.906 | ENSCANG00000038190 | AKAP8 | 100 | 88.759 | Colobus_angolensis_palliatus |
| ENSG00000105127 | AKAP8 | 100 | 77.874 | ENSCGRG00001016207 | Akap8 | 100 | 77.874 | Cricetulus_griseus_chok1gshd |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSCGRG00001019147 | - | 95 | 33.430 | Cricetulus_griseus_chok1gshd |
| ENSG00000105127 | AKAP8 | 84 | 32.805 | ENSCGRG00001000962 | - | 91 | 33.178 | Cricetulus_griseus_chok1gshd |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSCGRG00000010607 | - | 95 | 33.430 | Cricetulus_griseus_crigri |
| ENSG00000105127 | AKAP8 | 84 | 32.805 | ENSCGRG00000019338 | - | 91 | 33.178 | Cricetulus_griseus_crigri |
| ENSG00000105127 | AKAP8 | 100 | 77.066 | ENSCGRG00000010539 | Akap8 | 100 | 77.208 | Cricetulus_griseus_crigri |
| ENSG00000105127 | AKAP8 | 55 | 33.586 | ENSDARG00000099635 | akap8l | 91 | 30.435 | Danio_rerio |
| ENSG00000105127 | AKAP8 | 92 | 74.961 | ENSDNOG00000016300 | - | 99 | 72.429 | Dasypus_novemcinctus |
| ENSG00000105127 | AKAP8 | 95 | 68.639 | ENSDNOG00000036136 | - | 99 | 67.465 | Dasypus_novemcinctus |
| ENSG00000105127 | AKAP8 | 78 | 35.763 | ENSDNOG00000025063 | AKAP8L | 85 | 34.910 | Dasypus_novemcinctus |
| ENSG00000105127 | AKAP8 | 51 | 82.670 | ENSDORG00000027410 | - | 97 | 78.325 | Dipodomys_ordii |
| ENSG00000105127 | AKAP8 | 96 | 65.143 | ENSETEG00000013772 | - | 88 | 77.008 | Echinops_telfairi |
| ENSG00000105127 | AKAP8 | 68 | 34.051 | ENSEASG00005003631 | AKAP8L | 81 | 33.447 | Equus_asinus_asinus |
| ENSG00000105127 | AKAP8 | 99 | 78.161 | ENSEASG00005003609 | AKAP8 | 99 | 79.714 | Equus_asinus_asinus |
| ENSG00000105127 | AKAP8 | 66 | 34.615 | ENSECAG00000023532 | AKAP8L | 84 | 32.759 | Equus_caballus |
| ENSG00000105127 | AKAP8 | 100 | 83.595 | ENSECAG00000015084 | AKAP8 | 100 | 83.738 | Equus_caballus |
| ENSG00000105127 | AKAP8 | 55 | 36.250 | ENSELUG00000012410 | akap8l | 92 | 31.860 | Esox_lucius |
| ENSG00000105127 | AKAP8 | 66 | 34.879 | ENSFCAG00000034698 | AKAP8L | 87 | 34.520 | Felis_catus |
| ENSG00000105127 | AKAP8 | 92 | 83.515 | ENSFCAG00000013646 | AKAP8 | 100 | 78.621 | Felis_catus |
| ENSG00000105127 | AKAP8 | 69 | 35.577 | ENSFDAG00000020572 | AKAP8L | 92 | 33.997 | Fukomys_damarensis |
| ENSG00000105127 | AKAP8 | 99 | 82.653 | ENSFDAG00000008508 | AKAP8 | 99 | 82.799 | Fukomys_damarensis |
| ENSG00000105127 | AKAP8 | 77 | 58.333 | ENSGALG00000039773 | AKAP8 | 74 | 55.106 | Gallus_gallus |
| ENSG00000105127 | AKAP8 | 79 | 35.959 | ENSGALG00000039974 | AKAP8L | 75 | 35.582 | Gallus_gallus |
| ENSG00000105127 | AKAP8 | 91 | 53.550 | ENSGAGG00000025280 | AKAP8 | 90 | 52.883 | Gopherus_agassizii |
| ENSG00000105127 | AKAP8 | 78 | 36.667 | ENSGAGG00000025278 | AKAP8L | 80 | 36.860 | Gopherus_agassizii |
| ENSG00000105127 | AKAP8 | 100 | 97.977 | ENSGGOG00000013427 | AKAP8 | 100 | 97.977 | Gorilla_gorilla |
| ENSG00000105127 | AKAP8 | 66 | 36.895 | ENSGGOG00000013441 | AKAP8L | 95 | 32.216 | Gorilla_gorilla |
| ENSG00000105127 | AKAP8 | 100 | 82.857 | ENSHGLG00000006879 | AKAP8 | 100 | 83.000 | Heterocephalus_glaber_female |
| ENSG00000105127 | AKAP8 | 66 | 34.791 | ENSHGLG00000019540 | AKAP8L | 95 | 32.801 | Heterocephalus_glaber_female |
| ENSG00000105127 | AKAP8 | 66 | 34.791 | ENSHGLG00100007107 | AKAP8L | 95 | 32.801 | Heterocephalus_glaber_male |
| ENSG00000105127 | AKAP8 | 100 | 83.815 | ENSHGLG00100010349 | AKAP8 | 100 | 83.960 | Heterocephalus_glaber_male |
| ENSG00000105127 | AKAP8 | 50 | 39.385 | ENSIPUG00000012082 | akap8l | 90 | 33.662 | Ictalurus_punctatus |
| ENSG00000105127 | AKAP8 | 100 | 84.249 | ENSSTOG00000006762 | AKAP8 | 100 | 84.249 | Ictidomys_tridecemlineatus |
| ENSG00000105127 | AKAP8 | 66 | 35.354 | ENSSTOG00000022788 | AKAP8L | 95 | 33.043 | Ictidomys_tridecemlineatus |
| ENSG00000105127 | AKAP8 | 66 | 37.074 | ENSJJAG00000012334 | Akap8l | 96 | 32.994 | Jaculus_jaculus |
| ENSG00000105127 | AKAP8 | 99 | 75.543 | ENSJJAG00000011639 | Akap8 | 99 | 76.411 | Jaculus_jaculus |
| ENSG00000105127 | AKAP8 | 52 | 35.484 | ENSLBEG00000010793 | akap8l | 89 | 30.015 | Labrus_bergylta |
| ENSG00000105127 | AKAP8 | 91 | 43.605 | ENSLACG00000009083 | AKAP8 | 98 | 56.807 | Latimeria_chalumnae |
| ENSG00000105127 | AKAP8 | 89 | 34.384 | ENSLACG00000009932 | AKAP8L | 97 | 35.392 | Latimeria_chalumnae |
| ENSG00000105127 | AKAP8 | 79 | 34.673 | ENSLAFG00000004599 | AKAP8L | 90 | 35.549 | Loxodonta_africana |
| ENSG00000105127 | AKAP8 | 100 | 76.803 | ENSLAFG00000026120 | AKAP8 | 100 | 76.781 | Loxodonta_africana |
| ENSG00000105127 | AKAP8 | 100 | 96.098 | ENSMFAG00000042432 | AKAP8 | 100 | 96.098 | Macaca_fascicularis |
| ENSG00000105127 | AKAP8 | 66 | 36.768 | ENSMFAG00000045237 | AKAP8L | 81 | 34.589 | Macaca_fascicularis |
| ENSG00000105127 | AKAP8 | 99 | 95.918 | ENSMMUG00000017984 | AKAP8L | 100 | 95.918 | Macaca_mulatta |
| ENSG00000105127 | AKAP8 | 100 | 94.239 | ENSMNEG00000032407 | AKAP8 | 84 | 73.408 | Macaca_nemestrina |
| ENSG00000105127 | AKAP8 | 66 | 36.768 | ENSMNEG00000043813 | AKAP8L | 81 | 34.589 | Macaca_nemestrina |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSMLEG00000035928 | AKAP8L | 89 | 34.075 | Mandrillus_leucophaeus |
| ENSG00000105127 | AKAP8 | 99 | 95.912 | ENSMLEG00000034735 | AKAP8 | 100 | 95.912 | Mandrillus_leucophaeus |
| ENSG00000105127 | AKAP8 | 79 | 53.226 | ENSMGAG00000009065 | AKAP8 | 76 | 74.539 | Meleagris_gallopavo |
| ENSG00000105127 | AKAP8 | 75 | 35.575 | ENSMGAG00000009063 | AKAP8L | 95 | 35.563 | Meleagris_gallopavo |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSMAUG00000021519 | Akap8l | 81 | 33.677 | Mesocricetus_auratus |
| ENSG00000105127 | AKAP8 | 96 | 79.532 | ENSMAUG00000021975 | Akap8 | 100 | 78.653 | Mesocricetus_auratus |
| ENSG00000105127 | AKAP8 | 66 | 35.872 | ENSMICG00000027945 | AKAP8L | 88 | 33.230 | Microcebus_murinus |
| ENSG00000105127 | AKAP8 | 96 | 86.857 | ENSMICG00000012702 | AKAP8 | 100 | 80.460 | Microcebus_murinus |
| ENSG00000105127 | AKAP8 | 100 | 79.741 | ENSMOCG00000001588 | Akap8 | 100 | 79.741 | Microtus_ochrogaster |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSMOCG00000017098 | Akap8l | 95 | 34.055 | Microtus_ochrogaster |
| ENSG00000105127 | AKAP8 | 93 | 32.425 | ENSMODG00000014581 | AKAP8L | 85 | 33.814 | Monodelphis_domestica |
| ENSG00000105127 | AKAP8 | 100 | 62.771 | ENSMODG00000014573 | AKAP8 | 100 | 62.410 | Monodelphis_domestica |
| ENSG00000105127 | AKAP8 | 100 | 79.395 | MGP_CAROLIEiJ_G0021380 | Akap8 | 100 | 79.971 | Mus_caroli |
| ENSG00000105127 | AKAP8 | 66 | 35.772 | MGP_CAROLIEiJ_G0021381 | Akap8l | 81 | 34.359 | Mus_caroli |
| ENSG00000105127 | AKAP8 | 100 | 77.778 | ENSMUSG00000024045 | Akap8 | 100 | 79.076 | Mus_musculus |
| ENSG00000105127 | AKAP8 | 66 | 35.976 | ENSMUSG00000002625 | Akap8l | 81 | 34.359 | Mus_musculus |
| ENSG00000105127 | AKAP8 | 66 | 35.700 | MGP_PahariEiJ_G0020388 | Akap8l | 81 | 34.021 | Mus_pahari |
| ENSG00000105127 | AKAP8 | 100 | 78.561 | MGP_PahariEiJ_G0020387 | Akap8 | 100 | 78.561 | Mus_pahari |
| ENSG00000105127 | AKAP8 | 66 | 35.976 | MGP_SPRETEiJ_G0022295 | Akap8l | 88 | 34.059 | Mus_spretus |
| ENSG00000105127 | AKAP8 | 100 | 78.355 | MGP_SPRETEiJ_G0022294 | Akap8 | 100 | 79.971 | Mus_spretus |
| ENSG00000105127 | AKAP8 | 66 | 34.827 | ENSMPUG00000016078 | AKAP8L | 81 | 34.066 | Mustela_putorius_furo |
| ENSG00000105127 | AKAP8 | 100 | 82.615 | ENSMPUG00000016081 | AKAP8 | 100 | 83.333 | Mustela_putorius_furo |
| ENSG00000105127 | AKAP8 | 96 | 77.841 | ENSMLUG00000005460 | - | 99 | 84.517 | Myotis_lucifugus |
| ENSG00000105127 | AKAP8 | 66 | 35.800 | ENSMLUG00000005469 | AKAP8L | 81 | 33.503 | Myotis_lucifugus |
| ENSG00000105127 | AKAP8 | 100 | 81.609 | ENSNGAG00000010875 | Akap8 | 100 | 81.609 | Nannospalax_galili |
| ENSG00000105127 | AKAP8 | 100 | 97.110 | ENSNLEG00000005263 | AKAP8 | 100 | 97.110 | Nomascus_leucogenys |
| ENSG00000105127 | AKAP8 | 66 | 36.694 | ENSNLEG00000005271 | AKAP8L | 95 | 32.612 | Nomascus_leucogenys |
| ENSG00000105127 | AKAP8 | 56 | 73.333 | ENSMEUG00000008595 | - | 63 | 73.333 | Notamacropus_eugenii |
| ENSG00000105127 | AKAP8 | 58 | 37.037 | ENSMEUG00000014174 | AKAP8L | 95 | 31.376 | Notamacropus_eugenii |
| ENSG00000105127 | AKAP8 | 79 | 73.786 | ENSOPRG00000013430 | AKAP8 | 84 | 73.786 | Ochotona_princeps |
| ENSG00000105127 | AKAP8 | 99 | 82.267 | ENSODEG00000011314 | AKAP8 | 99 | 82.994 | Octodon_degus |
| ENSG00000105127 | AKAP8 | 78 | 33.162 | ENSODEG00000010840 | AKAP8L | 95 | 33.000 | Octodon_degus |
| ENSG00000105127 | AKAP8 | 100 | 73.496 | ENSOCUG00000004051 | - | 98 | 75.074 | Oryctolagus_cuniculus |
| ENSG00000105127 | AKAP8 | 80 | 53.571 | ENSOCUG00000027694 | - | 100 | 54.374 | Oryctolagus_cuniculus |
| ENSG00000105127 | AKAP8 | 65 | 36.235 | ENSOCUG00000011417 | AKAP8L | 85 | 34.901 | Oryctolagus_cuniculus |
| ENSG00000105127 | AKAP8 | 99 | 86.735 | ENSOGAG00000029564 | AKAP8 | 100 | 87.464 | Otolemur_garnettii |
| ENSG00000105127 | AKAP8 | 66 | 35.471 | ENSOGAG00000010943 | AKAP8L | 85 | 34.029 | Otolemur_garnettii |
| ENSG00000105127 | AKAP8 | 99 | 78.696 | ENSOARG00000003568 | AKAP8 | 99 | 79.046 | Ovis_aries |
| ENSG00000105127 | AKAP8 | 78 | 34.418 | ENSOARG00000003411 | AKAP8L | 88 | 34.069 | Ovis_aries |
| ENSG00000105127 | AKAP8 | 100 | 99.422 | ENSPPAG00000035505 | AKAP8 | 100 | 99.422 | Pan_paniscus |
| ENSG00000105127 | AKAP8 | 66 | 37.097 | ENSPPAG00000029573 | AKAP8L | 95 | 32.706 | Pan_paniscus |
| ENSG00000105127 | AKAP8 | 92 | 86.250 | ENSPPRG00000010081 | AKAP8 | 100 | 82.102 | Panthera_pardus |
| ENSG00000105127 | AKAP8 | 66 | 34.879 | ENSPPRG00000009539 | AKAP8L | 87 | 34.520 | Panthera_pardus |
| ENSG00000105127 | AKAP8 | 66 | 34.476 | ENSPTIG00000011001 | AKAP8L | 87 | 33.909 | Panthera_tigris_altaica |
| ENSG00000105127 | AKAP8 | 100 | 81.897 | ENSPTIG00000010640 | AKAP8 | 100 | 81.740 | Panthera_tigris_altaica |
| ENSG00000105127 | AKAP8 | 100 | 99.422 | ENSPTRG00000010620 | AKAP8 | 100 | 99.422 | Pan_troglodytes |
| ENSG00000105127 | AKAP8 | 66 | 37.097 | ENSPTRG00000010621 | AKAP8L | 95 | 32.706 | Pan_troglodytes |
| ENSG00000105127 | AKAP8 | 100 | 95.087 | ENSPANG00000015626 | AKAP8 | 100 | 95.087 | Papio_anubis |
| ENSG00000105127 | AKAP8 | 66 | 36.566 | ENSPANG00000009516 | AKAP8L | 81 | 34.589 | Papio_anubis |
| ENSG00000105127 | AKAP8 | 80 | 31.169 | ENSPSIG00000007626 | AKAP8L | 96 | 36.923 | Pelodiscus_sinensis |
| ENSG00000105127 | AKAP8 | 98 | 55.148 | ENSPSIG00000005989 | AKAP8 | 99 | 53.463 | Pelodiscus_sinensis |
| ENSG00000105127 | AKAP8 | 100 | 80.460 | ENSPEMG00000016622 | Akap8 | 100 | 80.460 | Peromyscus_maniculatus_bairdii |
| ENSG00000105127 | AKAP8 | 66 | 35.542 | ENSPEMG00000008156 | Akap8l | 95 | 32.851 | Peromyscus_maniculatus_bairdii |
| ENSG00000105127 | AKAP8 | 66 | 34.930 | ENSPCIG00000004037 | AKAP8L | 92 | 32.143 | Phascolarctos_cinereus |
| ENSG00000105127 | AKAP8 | 100 | 61.714 | ENSPCIG00000004006 | AKAP8 | 97 | 61.714 | Phascolarctos_cinereus |
| ENSG00000105127 | AKAP8 | 65 | 35.569 | ENSPPYG00000029684 | AKAP8L | 99 | 32.883 | Pongo_abelii |
| ENSG00000105127 | AKAP8 | 100 | 97.254 | ENSPPYG00000009671 | AKAP8 | 100 | 97.254 | Pongo_abelii |
| ENSG00000105127 | AKAP8 | 78 | 33.672 | ENSPCAG00000003928 | AKAP8L | 69 | 51.829 | Procavia_capensis |
| ENSG00000105127 | AKAP8 | 100 | 72.779 | ENSPCAG00000003834 | AKAP8 | 100 | 75.215 | Procavia_capensis |
| ENSG00000105127 | AKAP8 | 100 | 87.032 | ENSPCOG00000016236 | AKAP8 | 100 | 87.464 | Propithecus_coquereli |
| ENSG00000105127 | AKAP8 | 66 | 35.282 | ENSPCOG00000015753 | AKAP8L | 88 | 33.540 | Propithecus_coquereli |
| ENSG00000105127 | AKAP8 | 66 | 36.072 | ENSPVAG00000016917 | AKAP8L | 85 | 33.442 | Pteropus_vampyrus |
| ENSG00000105127 | AKAP8 | 100 | 83.405 | ENSPVAG00000016915 | AKAP8 | 100 | 84.704 | Pteropus_vampyrus |
| ENSG00000105127 | AKAP8 | 100 | 78.355 | ENSRNOG00000006559 | Akap8 | 100 | 79.683 | Rattus_norvegicus |
| ENSG00000105127 | AKAP8 | 66 | 35.944 | ENSRNOG00000006355 | Akap8l | 91 | 33.231 | Rattus_norvegicus |
| ENSG00000105127 | AKAP8 | 96 | 96.491 | ENSRBIG00000030066 | AKAP8 | 100 | 89.306 | Rhinopithecus_bieti |
| ENSG00000105127 | AKAP8 | 66 | 36.821 | ENSRBIG00000033532 | AKAP8L | 99 | 33.190 | Rhinopithecus_bieti |
| ENSG00000105127 | AKAP8 | 100 | 95.520 | ENSRROG00000031442 | AKAP8 | 100 | 95.520 | Rhinopithecus_roxellana |
| ENSG00000105127 | AKAP8 | 66 | 36.821 | ENSRROG00000031937 | AKAP8L | 95 | 33.286 | Rhinopithecus_roxellana |
| ENSG00000105127 | AKAP8 | 100 | 89.306 | ENSSBOG00000022494 | AKAP8 | 100 | 89.451 | Saimiri_boliviensis_boliviensis |
| ENSG00000105127 | AKAP8 | 67 | 36.634 | ENSSBOG00000031029 | AKAP8L | 95 | 33.047 | Saimiri_boliviensis_boliviensis |
| ENSG00000105127 | AKAP8 | 78 | 34.064 | ENSSHAG00000010686 | AKAP8L | 80 | 33.841 | Sarcophilus_harrisii |
| ENSG00000105127 | AKAP8 | 66 | 42.982 | ENSSFOG00015005929 | - | 79 | 42.241 | Scleropages_formosus |
| ENSG00000105127 | AKAP8 | 86 | 30.229 | ENSSFOG00015023854 | - | 95 | 30.435 | Scleropages_formosus |
| ENSG00000105127 | AKAP8 | 53 | 34.625 | ENSSMAG00000021221 | akap8l | 85 | 30.511 | Scophthalmus_maximus |
| ENSG00000105127 | AKAP8 | 56 | 34.848 | ENSSDUG00000013650 | akap8l | 90 | 31.445 | Seriola_dumerili |
| ENSG00000105127 | AKAP8 | 54 | 34.606 | ENSSLDG00000012237 | akap8l | 79 | 33.111 | Seriola_lalandi_dorsalis |
| ENSG00000105127 | AKAP8 | 96 | 75.439 | ENSSARG00000010714 | AKAP8 | 100 | 74.222 | Sorex_araneus |
| ENSG00000105127 | AKAP8 | 65 | 38.429 | ENSSPUG00000000622 | AKAP8L | 69 | 37.757 | Sphenodon_punctatus |
| ENSG00000105127 | AKAP8 | 51 | 35.000 | ENSSPAG00000018628 | akap8l | 86 | 30.551 | Stegastes_partitus |
| ENSG00000105127 | AKAP8 | 100 | 84.870 | ENSSSCG00000013837 | AKAP8 | 100 | 84.604 | Sus_scrofa |
| ENSG00000105127 | AKAP8 | 72 | 34.630 | ENSSSCG00000013838 | AKAP8L | 81 | 35.512 | Sus_scrofa |
| ENSG00000105127 | AKAP8 | 55 | 35.589 | ENSTRUG00000016038 | akap8l | 91 | 31.024 | Takifugu_rubripes |
| ENSG00000105127 | AKAP8 | 75 | 35.242 | ENSTTRG00000016002 | AKAP8L | 86 | 33.984 | Tursiops_truncatus |
| ENSG00000105127 | AKAP8 | 99 | 83.261 | ENSTTRG00000015996 | AKAP8 | 100 | 84.279 | Tursiops_truncatus |
| ENSG00000105127 | AKAP8 | 66 | 35.412 | ENSUAMG00000000463 | AKAP8L | 81 | 34.295 | Ursus_americanus |
| ENSG00000105127 | AKAP8 | 99 | 79.275 | ENSUAMG00000000693 | AKAP8 | 99 | 79.275 | Ursus_americanus |
| ENSG00000105127 | AKAP8 | 66 | 35.484 | ENSUMAG00000001892 | AKAP8L | 81 | 32.628 | Ursus_maritimus |
| ENSG00000105127 | AKAP8 | 100 | 79.971 | ENSUMAG00000001928 | AKAP8 | 100 | 79.971 | Ursus_maritimus |
| ENSG00000105127 | AKAP8 | 66 | 35.944 | ENSVVUG00000020394 | AKAP8L | 88 | 33.798 | Vulpes_vulpes |
| ENSG00000105127 | AKAP8 | 99 | 77.507 | ENSVVUG00000020457 | AKAP8 | 99 | 77.364 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000278 | mitotic cell cycle | 9473338. | TAS | Process |
| GO:0000793 | condensed chromosome | - | IEA | Component |
| GO:0001939 | female pronucleus | - | IEA | Component |
| GO:0003690 | double-stranded DNA binding | - | IEA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 9473338.12740381.16227597.16980585.22130794.23995757. | IPI | Function |
| GO:0005634 | nucleus | 9473338. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005794 | Golgi apparatus | - | IEA | Component |
| GO:0007076 | mitotic chromosome condensation | - | IEA | Process |
| GO:0007165 | signal transduction | 9473338. | TAS | Process |
| GO:0008270 | zinc ion binding | - | IEA | Function |
| GO:0015031 | protein transport | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016363 | nuclear matrix | 21873635. | IBA | Component |
| GO:0016363 | nuclear matrix | - | ISS | Component |
| GO:0031065 | positive regulation of histone deacetylation | 16980585. | IMP | Process |
| GO:0032720 | negative regulation of tumor necrosis factor production | - | IEA | Process |
| GO:0033127 | regulation of histone phosphorylation | 16980585. | IMP | Process |
| GO:0034237 | protein kinase A regulatory subunit binding | 21873635. | IBA | Function |
| GO:0034237 | protein kinase A regulatory subunit binding | 17911601. | IPI | Function |
| GO:0042826 | histone deacetylase binding | 16980585. | IDA | Function |
| GO:0044839 | cell cycle G2/M phase transition | 16980585. | IMP | Process |
| GO:0045087 | innate immune response | - | IEA | Process |
| GO:0051059 | NF-kappaB binding | - | IEA | Function |
| GO:0071222 | cellular response to lipopolysaccharide | - | IEA | Process |
| GO:0071380 | cellular response to prostaglandin E stimulus | - | IEA | Process |