
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000478946 | SAP | PF02037.27 | 2.4e-10 | 1 | 1 |
| ENSP00000484226 | SAP | PF02037.27 | 2.4e-10 | 1 | 1 |
| ENSP00000470687 | SAP | PF02037.27 | 1.2e-09 | 1 | 1 |
| ENSP00000375863 | SAP | PF02037.27 | 1.4e-09 | 1 | 1 |
| ENSP00000367460 | SAP | PF02037.27 | 2.9e-09 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000352456 | HNRNPUL1-202 | 3426 | XM_005258463 | ENSP00000340857 | 766 (aa) | XP_005258520 | A0A0A0MRA5 |
| ENST00000378215 | HNRNPUL1-203 | 2864 | - | ENSP00000367460 | 752 (aa) | - | Q9BUJ2 |
| ENST00000593587 | HNRNPUL1-205 | 2725 | - | ENSP00000472629 | 756 (aa) | - | Q9BUJ2 |
| ENST00000597725 | HNRNPUL1-211 | 573 | - | ENSP00000469858 | 172 (aa) | - | M0QYI8 |
| ENST00000600596 | HNRNPUL1-217 | 581 | - | ENSP00000469934 | 90 (aa) | - | M0QYM5 |
| ENST00000392006 | HNRNPUL1-204 | 3555 | XM_005258459 | ENSP00000375863 | 856 (aa) | XP_005258516 | Q9BUJ2 |
| ENST00000602130 | HNRNPUL1-220 | 2932 | - | ENSP00000470687 | 804 (aa) | - | Q9BUJ2 |
| ENST00000601309 | HNRNPUL1-218 | 672 | - | ENSP00000470793 | 118 (aa) | - | M0QZV6 |
| ENST00000599521 | HNRNPUL1-212 | 336 | - | - | - (aa) | - | - |
| ENST00000617774 | HNRNPUL1-222 | 451 | - | ENSP00000478946 | 122 (aa) | - | A0A087X1I2 |
| ENST00000600493 | HNRNPUL1-216 | 531 | - | - | - (aa) | - | - |
| ENST00000595806 | HNRNPUL1-210 | 1659 | - | - | - (aa) | - | - |
| ENST00000595196 | HNRNPUL1-208 | 3043 | - | ENSP00000472315 | 105 (aa) | - | M0R247 |
| ENST00000599614 | HNRNPUL1-213 | 2447 | - | ENSP00000473178 | 641 (aa) | - | M0R3F1 |
| ENST00000594207 | HNRNPUL1-206 | 816 | - | - | - (aa) | - | - |
| ENST00000601336 | HNRNPUL1-219 | 754 | - | ENSP00000471302 | 158 (aa) | - | M0R0K8 |
| ENST00000595018 | HNRNPUL1-207 | 2794 | - | ENSP00000473132 | 756 (aa) | - | Q9BUJ2 |
| ENST00000599719 | HNRNPUL1-214 | 1473 | - | ENSP00000470172 | 371 (aa) | - | M0QYZ0 |
| ENST00000595336 | HNRNPUL1-209 | 393 | - | ENSP00000472221 | 81 (aa) | - | M0R203 |
| ENST00000617305 | HNRNPUL1-221 | 1018 | - | ENSP00000484226 | 122 (aa) | - | A0A087X1I2 |
| ENST00000600332 | HNRNPUL1-215 | 644 | - | ENSP00000469816 | 26 (aa) | - | M0QYG9 |
| ENST00000263367 | HNRNPUL1-201 | 2952 | XM_005258461 | ENSP00000263367 | 767 (aa) | XP_005258518 | B7Z4B8 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05164 | Influenza A | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000105323 | rs13345456 | 19 | 41301013 | C | Heel bone mineral density | 28869591 | [0.018-0.033] unit decrease | 0.0255354 | EFO_0009270 |
| ENSG00000105323 | rs13345456 | 19 | 41301013 | C | Heel bone mineral density | 28869591 | [0.023-0.044] unit decrease | 0.0337348 | EFO_0009270 |
| ENSG00000105323 | rs746549124 | 19 | 41278449 | ? | Heel bone mineral density | 30048462 | [0.018-0.028] unit decrease | 0.0229806 | EFO_0009270 |
| ENSG00000105323 | rs13345456 | 19 | 41301013 | ? | Heel bone mineral density | 30048462 | [0.015-0.024] unit decrease | 0.0194043 | EFO_0009270 |
| ENSG00000105323 | rs12983339 | 19 | 41264985 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 |
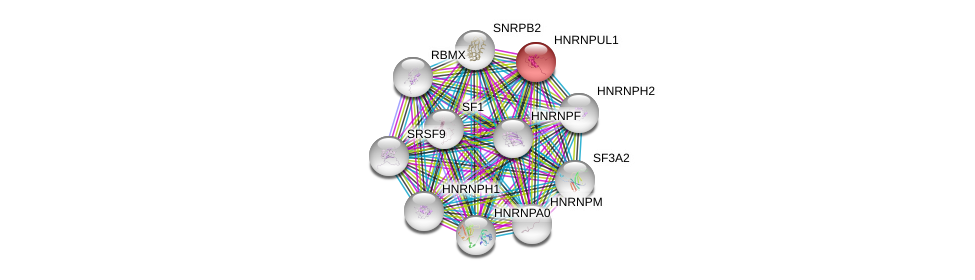
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000105323 | HNRNPUL1 | 98 | 50.000 | ENSG00000214753 | HNRNPUL2 | 86 | 43.981 |
| ENSG00000105323 | HNRNPUL1 | 98 | 60.227 | ENSG00000153187 | HNRNPU | 99 | 59.000 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSAPOG00000013078 | HNRNPUL1 | 67 | 78.058 | Acanthochromis_polyacanthus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.547 | ENSAMEG00000011681 | HNRNPUL1 | 100 | 98.133 | Ailuropoda_melanoleuca |
| ENSG00000105323 | HNRNPUL1 | 98 | 86.364 | ENSACIG00000024072 | HNRNPUL1 | 67 | 78.777 | Amphilophus_citrinellus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSAOCG00000006694 | HNRNPUL1 | 57 | 81.250 | Amphiprion_ocellaris |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSAPEG00000014298 | HNRNPUL1 | 57 | 81.250 | Amphiprion_percula |
| ENSG00000105323 | HNRNPUL1 | 98 | 81.818 | ENSATEG00000008814 | HNRNPUL1 | 77 | 60.320 | Anabas_testudineus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSANAG00000027262 | HNRNPUL1 | 100 | 99.069 | Aotus_nancymaae |
| ENSG00000105323 | HNRNPUL1 | 98 | 87.500 | ENSAMXG00000004623 | - | 66 | 78.417 | Astyanax_mexicanus |
| ENSG00000105323 | HNRNPUL1 | 98 | 86.364 | ENSAMXG00000013644 | HNRNPUL1 | 77 | 63.798 | Astyanax_mexicanus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.256 | ENSBTAG00000003168 | HNRNPUL1 | 100 | 97.669 | Bos_taurus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSCJAG00000020106 | HNRNPUL1 | 100 | 97.575 | Callithrix_jacchus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.942 | ENSCAFG00000005026 | HNRNPUL1 | 100 | 98.481 | Canis_familiaris |
| ENSG00000105323 | HNRNPUL1 | 97 | 100.000 | ENSCAFG00020014782 | HNRNPUL1 | 100 | 94.412 | Canis_lupus_dingo |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.256 | ENSCHIG00000015165 | HNRNPUL1 | 100 | 94.289 | Capra_hircus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSCPOG00000008101 | - | 100 | 95.333 | Cavia_porcellus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSCCAG00000038049 | HNRNPUL1 | 100 | 97.806 | Cebus_capucinus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSCATG00000039807 | HNRNPUL1 | 100 | 99.734 | Cercocebus_atys |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSCLAG00000000985 | HNRNPUL1 | 100 | 96.966 | Chinchilla_lanigera |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSCSAG00000003544 | HNRNPUL1 | 100 | 99.650 | Chlorocebus_sabaeus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSCANG00000027995 | HNRNPUL1 | 100 | 99.601 | Colobus_angolensis_palliatus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSCGRG00001015368 | Hnrnpul1 | 87 | 93.841 | Cricetulus_griseus_chok1gshd |
| ENSG00000105323 | HNRNPUL1 | 98 | 78.409 | ENSCSEG00000005657 | HNRNPUL1 | 61 | 75.180 | Cynoglossus_semilaevis |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSCVAG00000009524 | - | 72 | 78.231 | Cyprinodon_variegatus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSDORG00000003603 | Hnrnpul1 | 92 | 92.193 | Dipodomys_ordii |
| ENSG00000105323 | HNRNPUL1 | 87 | 55.814 | ENSEBUG00000012163 | HNRNPUL1 | 70 | 78.218 | Eptatretus_burgeri |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.837 | ENSEASG00005006344 | HNRNPUL1 | 100 | 98.133 | Equus_asinus_asinus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.837 | ENSECAG00000024149 | HNRNPUL1 | 100 | 98.019 | Equus_caballus |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSELUG00000021668 | - | 51 | 75.983 | Esox_lucius |
| ENSG00000105323 | HNRNPUL1 | 98 | 87.500 | ENSELUG00000019520 | HNRNPUL1 | 61 | 75.000 | Esox_lucius |
| ENSG00000105323 | HNRNPUL1 | 100 | 99.419 | ENSFCAG00000005433 | HNRNPUL1 | 100 | 98.715 | Felis_catus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.256 | ENSFDAG00000011784 | HNRNPUL1 | 93 | 96.715 | Fukomys_damarensis |
| ENSG00000105323 | HNRNPUL1 | 98 | 82.955 | ENSGAFG00000015786 | HNRNPUL1 | 71 | 71.290 | Gambusia_affinis |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSGGOG00000009804 | HNRNPUL1 | 100 | 99.734 | Gorilla_gorilla |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSHBUG00000010129 | HNRNPUL1 | 77 | 61.493 | Haplochromis_burtoni |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.256 | ENSHGLG00000007171 | HNRNPUL1 | 100 | 96.733 | Heterocephalus_glaber_female |
| ENSG00000105323 | HNRNPUL1 | 100 | 90.349 | ENSHGLG00100005101 | HNRNPUL1 | 89 | 97.059 | Heterocephalus_glaber_male |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSHCOG00000017927 | HNRNPUL1 | 78 | 59.259 | Hippocampus_comes |
| ENSG00000105323 | HNRNPUL1 | 98 | 88.636 | ENSIPUG00000024012 | HNRNPUL1 | 67 | 64.632 | Ictalurus_punctatus |
| ENSG00000105323 | HNRNPUL1 | 99 | 54.146 | ENSIPUG00000011639 | - | 75 | 59.738 | Ictalurus_punctatus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.256 | ENSSTOG00000001605 | HNRNPUL1 | 100 | 97.550 | Ictidomys_tridecemlineatus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSJJAG00000011986 | Hnrnpul1 | 100 | 85.830 | Jaculus_jaculus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSKMAG00000009438 | HNRNPUL1 | 76 | 60.769 | Kryptolebias_marmoratus |
| ENSG00000105323 | HNRNPUL1 | 98 | 86.364 | ENSLBEG00000018092 | HNRNPUL1 | 69 | 74.820 | Labrus_bergylta |
| ENSG00000105323 | HNRNPUL1 | 98 | 94.318 | ENSLACG00000018039 | HNRNPUL1 | 68 | 77.104 | Latimeria_chalumnae |
| ENSG00000105323 | HNRNPUL1 | 98 | 88.636 | ENSLOCG00000014236 | HNRNPUL1 | 66 | 80.576 | Lepisosteus_oculatus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSMFAG00000034257 | HNRNPUL1 | 100 | 99.734 | Macaca_fascicularis |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSMMUG00000008139 | HNRNPUL1 | 100 | 99.734 | Macaca_mulatta |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSMNEG00000035264 | HNRNPUL1 | 100 | 99.734 | Macaca_nemestrina |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSMLEG00000024704 | HNRNPUL1 | 100 | 99.734 | Mandrillus_leucophaeus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSMAMG00000014383 | HNRNPUL1 | 78 | 63.287 | Mastacembelus_armatus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSMZEG00005024205 | HNRNPUL1 | 77 | 61.377 | Maylandia_zebra |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSMAUG00000020435 | Hnrnpul1 | 88 | 95.922 | Mesocricetus_auratus |
| ENSG00000105323 | HNRNPUL1 | 100 | 97.754 | ENSMICG00000001989 | HNRNPUL1 | 100 | 97.550 | Microcebus_murinus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSMOCG00000008200 | Hnrnpul1 | 88 | 95.752 | Microtus_ochrogaster |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSMMOG00000004808 | HNRNPUL1 | 74 | 63.666 | Mola_mola |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | MGP_CAROLIEiJ_G0029381 | Hnrnpul1 | 99 | 92.604 | Mus_caroli |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSMUSG00000040725 | Hnrnpul1 | 99 | 92.308 | Mus_musculus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | MGP_PahariEiJ_G0021234 | Hnrnpul1 | 99 | 92.012 | Mus_pahari |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | MGP_SPRETEiJ_G0030477 | Hnrnpul1 | 99 | 92.308 | Mus_spretus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.383 | ENSMPUG00000017654 | HNRNPUL1 | 94 | 97.664 | Mustela_putorius_furo |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSNGAG00000017197 | Hnrnpul1 | 88 | 96.574 | Nannospalax_galili |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSNBRG00000004398 | HNRNPUL1 | 77 | 62.826 | Neolamprologus_brichardi |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSNLEG00000013751 | HNRNPUL1 | 100 | 99.180 | Nomascus_leucogenys |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSOPRG00000008773 | HNRNPUL1 | 100 | 93.349 | Ochotona_princeps |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSODEG00000000913 | HNRNPUL1 | 100 | 96.499 | Octodon_degus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSONIG00000004004 | HNRNPUL1 | 67 | 78.777 | Oreochromis_niloticus |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSOCUG00000016666 | HNRNPUL1 | 100 | 96.383 | Oryctolagus_cuniculus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSORLG00000001002 | HNRNPUL1 | 80 | 62.090 | Oryzias_latipes |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSORLG00020020272 | HNRNPUL1 | 78 | 62.980 | Oryzias_latipes_hni |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSORLG00015009059 | HNRNPUL1 | 69 | 73.226 | Oryzias_latipes_hsok |
| ENSG00000105323 | HNRNPUL1 | 98 | 86.364 | ENSOMEG00000021915 | HNRNPUL1 | 75 | 63.014 | Oryzias_melastigma |
| ENSG00000105323 | HNRNPUL1 | 100 | 97.886 | ENSOGAG00000013395 | - | 100 | 97.433 | Otolemur_garnettii |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSPPAG00000038140 | HNRNPUL1 | 96 | 99.214 | Pan_paniscus |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.545 | ENSPPRG00000003397 | HNRNPUL1 | 100 | 98.248 | Panthera_pardus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSPTRG00000011019 | HNRNPUL1 | 100 | 99.867 | Pan_troglodytes |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSPANG00000009467 | HNRNPUL1 | 100 | 99.734 | Papio_anubis |
| ENSG00000105323 | HNRNPUL1 | 98 | 90.909 | ENSPKIG00000017308 | - | 72 | 79.496 | Paramormyrops_kingsleyae |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSPEMG00000009618 | Hnrnpul1 | 97 | 93.731 | Peromyscus_maniculatus_bairdii |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSPFOG00000014230 | HNRNPUL1 | 53 | 81.250 | Poecilia_formosa |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSPLAG00000004833 | HNRNPUL1 | 67 | 73.226 | Poecilia_latipinna |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSPREG00000017804 | HNRNPUL1 | 67 | 77.698 | Poecilia_reticulata |
| ENSG00000105323 | HNRNPUL1 | 100 | 99.367 | ENSPPYG00000010011 | HNRNPUL1 | 99 | 95.789 | Pongo_abelii |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSPCAG00000013414 | HNRNPUL1 | 92 | 94.108 | Procavia_capensis |
| ENSG00000105323 | HNRNPUL1 | 99 | 100.000 | ENSPCOG00000020358 | HNRNPUL1 | 100 | 96.886 | Propithecus_coquereli |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSPNYG00000007583 | HNRNPUL1 | 77 | 62.654 | Pundamilia_nyererei |
| ENSG00000105323 | HNRNPUL1 | 98 | 88.636 | ENSPNAG00000019775 | HNRNPUL1 | 80 | 63.677 | Pygocentrus_nattereri |
| ENSG00000105323 | HNRNPUL1 | 98 | 100.000 | ENSRNOG00000020683 | Hnrnpul1 | 88 | 96.078 | Rattus_norvegicus |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSRBIG00000036625 | HNRNPUL1 | 100 | 99.468 | Rhinopithecus_bieti |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSRROG00000030124 | HNRNPUL1 | 100 | 99.468 | Rhinopithecus_roxellana |
| ENSG00000105323 | HNRNPUL1 | 100 | 100.000 | ENSSBOG00000035512 | HNRNPUL1 | 100 | 99.065 | Saimiri_boliviensis_boliviensis |
| ENSG00000105323 | HNRNPUL1 | 98 | 92.045 | ENSSFOG00015022940 | HNRNPUL1 | 68 | 79.859 | Scleropages_formosus |
| ENSG00000105323 | HNRNPUL1 | 98 | 88.636 | ENSSFOG00015007000 | - | 83 | 63.691 | Scleropages_formosus |
| ENSG00000105323 | HNRNPUL1 | 98 | 87.500 | ENSSMAG00000020270 | HNRNPUL1 | 61 | 76.259 | Scophthalmus_maximus |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSSDUG00000000399 | HNRNPUL1 | 69 | 77.698 | Seriola_dumerili |
| ENSG00000105323 | HNRNPUL1 | 98 | 84.091 | ENSSLDG00000005140 | HNRNPUL1 | 59 | 76.044 | Seriola_lalandi_dorsalis |
| ENSG00000105323 | HNRNPUL1 | 89 | 90.000 | ENSSPUG00000014431 | HNRNPUL1 | 75 | 68.058 | Sphenodon_punctatus |
| ENSG00000105323 | HNRNPUL1 | 98 | 85.227 | ENSSPAG00000018378 | HNRNPUL1 | 77 | 61.152 | Stegastes_partitus |
| ENSG00000105323 | HNRNPUL1 | 98 | 79.545 | ENSTRUG00000015154 | HNRNPUL1 | 64 | 75.180 | Takifugu_rubripes |
| ENSG00000105323 | HNRNPUL1 | 100 | 98.942 | ENSVVUG00000005386 | HNRNPUL1 | 100 | 98.481 | Vulpes_vulpes |
| ENSG00000105323 | HNRNPUL1 | 98 | 81.818 | ENSXCOG00000012492 | HNRNPUL1 | 65 | 77.338 | Xiphophorus_couchianus |
| ENSG00000105323 | HNRNPUL1 | 98 | 81.818 | ENSXMAG00000004815 | HNRNPUL1 | 67 | 76.978 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 12489984.17267598.21516116.22365833.24965446.25416956. | IPI | Function |
| GO:0005634 | nucleus | 11513728. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0006396 | RNA processing | 9733834. | TAS | Process |
| GO:0009615 | response to virus | - | IEA | Process |
| GO:0019899 | enzyme binding | 11513728. | IPI | Function |