
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000439616 | OGDH-203 | 2962 | XM_024446783 | ENSP00000398576 | 873 (aa) | XP_024302551 | E9PFG7 |
| ENST00000443864 | OGDH-204 | 1744 | - | ENSP00000388084 | 427 (aa) | - | Q02218 |
| ENST00000449767 | OGDH-207 | 3479 | - | ENSP00000392878 | 1019 (aa) | - | Q02218 |
| ENST00000497326 | OGDH-209 | 554 | - | - | - (aa) | - | - |
| ENST00000459672 | OGDH-208 | 601 | - | - | - (aa) | - | - |
| ENST00000444676 | OGDH-205 | 3309 | XM_005249759 | ENSP00000414662 | 1038 (aa) | XP_005249816 | E9PCR7 |
| ENST00000631326 | OGDH-210 | 4116 | - | ENSP00000486854 | 1001 (aa) | - | A0A0D9SFS3 |
| ENST00000419661 | OGDH-202 | 789 | - | ENSP00000411830 | 211 (aa) | - | C9J4G7 |
| ENST00000222673 | OGDH-201 | 4181 | - | ENSP00000222673 | 1023 (aa) | - | Q02218 |
| ENST00000447398 | OGDH-206 | 3474 | XM_005249761 | ENSP00000388183 | 1034 (aa) | XP_005249818 | E9PDF2 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000105953 | rs17133935 | 7 | 44698080 | G | Systolic blood pressure change trajectories | 27980656 | [0.18-0.42] unit increase | 0.3 | EFO_0006944 |
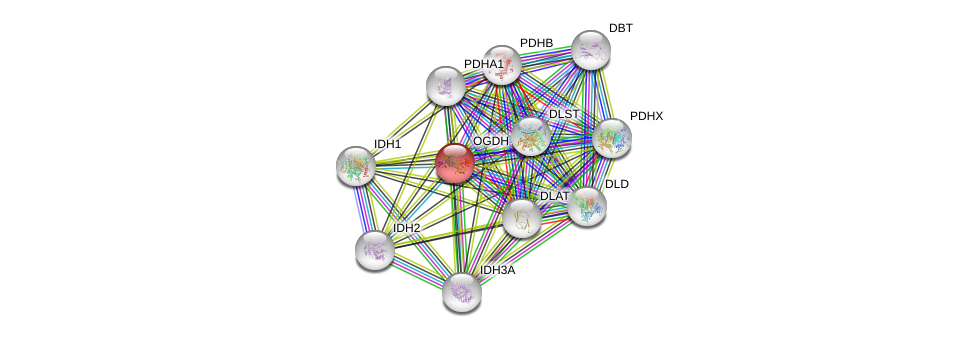
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000105953 | OGDH | 97 | 65.335 | FBgn0010352 | Nc73EF | 98 | 62.287 | Drosophila_melanogaster |
| ENSG00000105953 | OGDH | 100 | 96.209 | ENSMUSG00000020456 | Ogdh | 100 | 97.619 | Mus_musculus |
| ENSG00000105953 | OGDH | 99 | 45.507 | YIL125W | KGD1 | 99 | 45.507 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity | 21873635. | IBA | Function |
| GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity | 24495017.29211711. | IDA | Function |
| GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity | - | ISS | Function |
| GO:0005634 | nucleus | 29211711. | IDA | Component |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 29211711. | IDA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0006091 | generation of precursor metabolites and energy | - | ISS | Process |
| GO:0006096 | glycolytic process | - | IEA | Process |
| GO:0006099 | tricarboxylic acid cycle | 21873635. | IBA | Process |
| GO:0006099 | tricarboxylic acid cycle | - | TAS | Process |
| GO:0006103 | 2-oxoglutarate metabolic process | 24495017.29211711. | IDA | Process |
| GO:0006104 | succinyl-CoA metabolic process | 24495017.29211711. | IDA | Process |
| GO:0006554 | lysine catabolic process | - | TAS | Process |
| GO:0006734 | NADH metabolic process | - | IEA | Process |
| GO:0021695 | cerebellar cortex development | - | IEA | Process |
| GO:0021756 | striatum development | - | IEA | Process |
| GO:0021766 | hippocampus development | - | IEA | Process |
| GO:0021794 | thalamus development | - | IEA | Process |
| GO:0021860 | pyramidal neuron development | - | IEA | Process |
| GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb | - | IEA | Process |
| GO:0030976 | thiamine pyrophosphate binding | 24495017. | IDA | Function |
| GO:0031072 | heat shock protein binding | - | IEA | Function |
| GO:0031966 | mitochondrial membrane | - | ISS | Component |
| GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity | - | IEA | Function |
| GO:0045252 | oxoglutarate dehydrogenase complex | 21873635. | IBA | Component |
| GO:0045252 | oxoglutarate dehydrogenase complex | 29211711. | IDA | Component |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0051087 | chaperone binding | - | IEA | Function |
| GO:0061034 | olfactory bulb mitral cell layer development | - | IEA | Process |
| GO:0106077 | histone succinylation | 29211711. | IDA | Process |