
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000409143 | UvrD-helicase | PF00580.21 | 6.5e-08 | 1 | 1 |
| ENSP00000224140 | UvrD-helicase | PF00580.21 | 0.00028 | 1 | 1 |
| PID | Title | Method | Time | Author | Doi |
|---|---|---|---|---|---|
| 29431736 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000464133 | SETX-203 | 631 | - | - | - (aa) | - | - |
| ENST00000436441 | SETX-202 | 5730 | XM_017014496 | ENSP00000409143 | 948 (aa) | XP_016869985 | X6RI79 |
| ENST00000477049 | SETX-205 | 4067 | - | - | - (aa) | - | - |
| ENST00000474172 | SETX-204 | 719 | - | - | - (aa) | - | - |
| ENST00000224140 | SETX-201 | 11100 | XM_005272172 | ENSP00000224140 | 2677 (aa) | XP_005272229 | Q7Z333 |
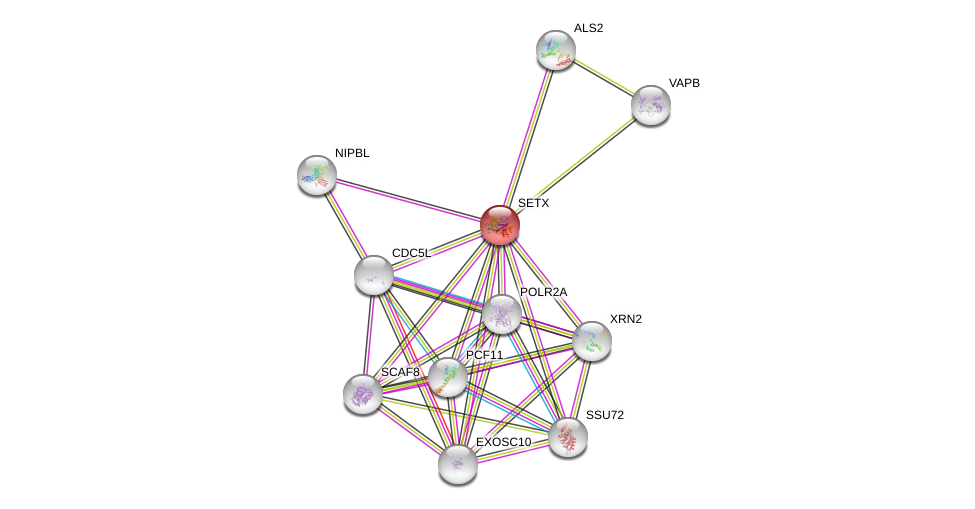
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000107290 | SETX | 76 | 44.054 | ENSAMXG00000014406 | - | 85 | 42.631 | Astyanax_mexicanus |
| ENSG00000107290 | SETX | 100 | 89.557 | ENSCJAG00000018245 | SETX | 100 | 89.258 | Callithrix_jacchus |
| ENSG00000107290 | SETX | 90 | 80.070 | ENSCAFG00020024489 | SETX | 100 | 82.091 | Canis_lupus_dingo |
| ENSG00000107290 | SETX | 84 | 80.605 | ENSTSYG00000030808 | - | 92 | 80.605 | Carlito_syrichta |
| ENSG00000107290 | SETX | 100 | 78.714 | ENSCPOG00000001492 | SETX | 100 | 71.561 | Cavia_porcellus |
| ENSG00000107290 | SETX | 99 | 79.325 | ENSCLAG00000012582 | SETX | 99 | 73.976 | Chinchilla_lanigera |
| ENSG00000107290 | SETX | 99 | 67.300 | ENSETEG00000008111 | - | 74 | 91.898 | Echinops_telfairi |
| ENSG00000107290 | SETX | 91 | 82.658 | ENSECAG00000012019 | SETX | 99 | 82.658 | Equus_caballus |
| ENSG00000107290 | SETX | 80 | 42.693 | ENSELUG00000022536 | - | 83 | 42.714 | Esox_lucius |
| ENSG00000107290 | SETX | 100 | 81.349 | ENSFCAG00000007102 | SETX | 100 | 77.749 | Felis_catus |
| ENSG00000107290 | SETX | 80 | 67.013 | ENSFALG00000001464 | SETX | 89 | 67.013 | Ficedula_albicollis |
| ENSG00000107290 | SETX | 100 | 97.273 | ENSGGOG00000027664 | SETX | 100 | 97.273 | Gorilla_gorilla |
| ENSG00000107290 | SETX | 79 | 44.778 | ENSIPUG00000006685 | setx | 85 | 45.008 | Ictalurus_punctatus |
| ENSG00000107290 | SETX | 83 | 55.593 | ENSLOCG00000004264 | SETX | 91 | 55.760 | Lepisosteus_oculatus |
| ENSG00000107290 | SETX | 71 | 91.207 | ENSLAFG00000017327 | - | 100 | 81.119 | Loxodonta_africana |
| ENSG00000107290 | SETX | 97 | 76.247 | ENSMOCG00000000848 | Setx | 99 | 68.000 | Microtus_ochrogaster |
| ENSG00000107290 | SETX | 100 | 78.864 | MGP_PahariEiJ_G0024821 | Setx | 100 | 72.080 | Mus_pahari |
| ENSG00000107290 | SETX | 99 | 93.150 | ENSNLEG00000012839 | SETX | 100 | 93.150 | Nomascus_leucogenys |
| ENSG00000107290 | SETX | 96 | 79.235 | ENSOCUG00000005616 | SETX | 96 | 71.528 | Oryctolagus_cuniculus |
| ENSG00000107290 | SETX | 99 | 85.338 | ENSOGAG00000013387 | SETX | 99 | 80.751 | Otolemur_garnettii |
| ENSG00000107290 | SETX | 100 | 98.245 | ENSPPAG00000004108 | SETX | 100 | 98.245 | Pan_paniscus |
| ENSG00000107290 | SETX | 100 | 81.560 | ENSPPRG00000012867 | SETX | 100 | 77.786 | Panthera_pardus |
| ENSG00000107290 | SETX | 100 | 81.560 | ENSPTIG00000021101 | SETX | 100 | 77.646 | Panthera_tigris_altaica |
| ENSG00000107290 | SETX | 100 | 98.282 | ENSPTRG00000021495 | SETX | 100 | 98.282 | Pan_troglodytes |
| ENSG00000107290 | SETX | 75 | 49.256 | ENSPKIG00000025162 | SETX | 84 | 49.587 | Paramormyrops_kingsleyae |
| ENSG00000107290 | SETX | 59 | 93.103 | ENSSARG00000002336 | - | 63 | 93.103 | Sorex_araneus |
| ENSG00000107290 | SETX | 88 | 66.519 | ENSSPUG00000016332 | SETX | 88 | 70.494 | Sphenodon_punctatus |
| ENSG00000107290 | SETX | 93 | 86.631 | ENSSSCG00000005724 | SETX | 99 | 76.198 | Sus_scrofa |
| ENSG00000107290 | SETX | 79 | 71.833 | ENSTGUG00000005117 | SETX | 86 | 71.833 | Taeniopygia_guttata |
| ENSG00000107290 | SETX | 84 | 86.507 | ENSUMAG00000016729 | SETX | 92 | 88.129 | Ursus_maritimus |
| ENSG00000107290 | SETX | 79 | 63.123 | ENSXETG00000006254 | setx | 95 | 63.411 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000165 | MAPK cascade | 21576111. | IDA | Process |
| GO:0000228 | nuclear chromosome | 24105744. | IDA | Component |
| GO:0000781 | chromosome, telomeric region | - | IEA | Component |
| GO:0001147 | transcription termination site sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0001147 | transcription termination site sequence-specific DNA binding | 21700224. | IDA | Function |
| GO:0003677 | DNA binding | 17562789. | IC | Function |
| GO:0003678 | DNA helicase activity | 17562789. | TAS | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 19515850.20936779.24105744.24244371.26700805. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 17562789.21576111. | IDA | Component |
| GO:0005654 | nucleoplasm | 17562789. | IDA | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005737 | cytoplasm | 17562789.21576111. | IDA | Component |
| GO:0006302 | double-strand break repair | 17562789. | IDA | Process |
| GO:0006310 | DNA recombination | - | IEA | Process |
| GO:0006353 | DNA-templated transcription, termination | 26700805. | IMP | Process |
| GO:0006369 | termination of RNA polymerase II transcription | 21873635. | IBA | Process |
| GO:0006376 | mRNA splice site selection | 19515850. | IMP | Process |
| GO:0006396 | RNA processing | 17562789. | TAS | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 17562789. | IDA | Process |
| GO:0007283 | spermatogenesis | - | IEA | Process |
| GO:0007623 | circadian rhythm | - | IEA | Process |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 21576111. | IDA | Process |
| GO:0010976 | positive regulation of neuron projection development | 21576111. | IDA | Process |
| GO:0016604 | nuclear body | 21873635. | IBA | Component |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0030424 | axon | 21576111. | IDA | Component |
| GO:0030426 | growth cone | 21576111. | IDA | Component |
| GO:0032508 | DNA duplex unwinding | - | IEA | Process |
| GO:0033120 | positive regulation of RNA splicing | 19515850. | IMP | Process |
| GO:0034599 | cellular response to oxidative stress | 17562789. | IDA | Process |
| GO:0042802 | identical protein binding | 24244371. | IPI | Function |
| GO:0043066 | negative regulation of apoptotic process | 21576111. | IDA | Process |
| GO:0043491 | protein kinase B signaling | 21576111. | IDA | Process |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | 21576111. | IDA | Process |
| GO:0045171 | intercellular bridge | - | IDA | Component |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 19515850. | IMP | Process |
| GO:0060566 | positive regulation of DNA-templated transcription, termination | 19515850. | IMP | Process |
| GO:0070301 | cellular response to hydrogen peroxide | 17562789. | IDA | Process |
| GO:0071300 | cellular response to retinoic acid | 21576111. | IDA | Process |
| GO:2000144 | positive regulation of DNA-templated transcription, initiation | 21700224. | IMP | Process |
| GO:2000806 | positive regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 21700224. | IMP | Process |