
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000531244 | PPP6R3-222 | 687 | - | ENSP00000432837 | 186 (aa) | - | E9PNN8 |
| ENST00000529172 | PPP6R3-216 | 500 | - | - | - (aa) | - | - |
| ENST00000528635 | PPP6R3-215 | 538 | - | ENSP00000436789 | 8 (aa) | - | A0A1D5RMU2 |
| ENST00000524845 | PPP6R3-205 | 2771 | - | ENSP00000431415 | 844 (aa) | - | Q5H9R7 |
| ENST00000524904 | PPP6R3-206 | 4292 | XM_006718612 | ENSP00000433058 | 867 (aa) | XP_006718675 | Q5H9R7 |
| ENST00000525152 | PPP6R3-208 | 582 | - | - | - (aa) | - | - |
| ENST00000393800 | PPP6R3-203 | 5093 | XM_006718608 | ENSP00000377389 | 873 (aa) | XP_006718671 | Q5H9R7 |
| ENST00000393801 | PPP6R3-204 | 5069 | XM_011545137 | ENSP00000377390 | 879 (aa) | XP_011543439 | Q5H9R7 |
| ENST00000526574 | PPP6R3-211 | 489 | - | - | - (aa) | - | - |
| ENST00000530427 | PPP6R3-220 | 560 | - | - | - (aa) | - | - |
| ENST00000534534 | PPP6R3-226 | 2968 | - | ENSP00000434429 | 641 (aa) | - | E9PQP7 |
| ENST00000527403 | PPP6R3-214 | 4862 | XM_017017974 | ENSP00000433565 | 838 (aa) | XP_016873463 | E9PKF6 |
| ENST00000533127 | PPP6R3-224 | 551 | - | ENSP00000433768 | 39 (aa) | - | E9PJD8 |
| ENST00000529907 | PPP6R3-219 | 826 | - | ENSP00000431738 | 140 (aa) | - | E9PK08 |
| ENST00000526307 | PPP6R3-210 | 4134 | - | - | - (aa) | - | - |
| ENST00000525421 | PPP6R3-209 | 830 | - | ENSP00000434426 | 112 (aa) | - | H0YDW1 |
| ENST00000525050 | PPP6R3-207 | 513 | - | - | - (aa) | - | - |
| ENST00000530734 | PPP6R3-221 | 576 | - | ENSP00000433853 | 51 (aa) | - | H0YDK9 |
| ENST00000265637 | PPP6R3-202 | 4783 | XM_006718621 | ENSP00000265637 | 827 (aa) | XP_006718684 | H7BXH2 |
| ENST00000534190 | PPP6R3-225 | 1980 | XM_024448603 | ENSP00000436209 | 580 (aa) | XP_024304371 | H0YEN2 |
| ENST00000526593 | PPP6R3-212 | 570 | - | ENSP00000436601 | 109 (aa) | - | H0YEV0 |
| ENST00000529710 | PPP6R3-218 | 3228 | - | ENSP00000437329 | 791 (aa) | - | Q5H9R7 |
| ENST00000529344 | PPP6R3-217 | 603 | - | ENSP00000433551 | 75 (aa) | - | E9PKG4 |
| ENST00000527069 | PPP6R3-213 | 573 | - | - | - (aa) | - | - |
| ENST00000531432 | PPP6R3-223 | 589 | - | - | - (aa) | - | - |
| ENST00000265636 | PPP6R3-201 | 4794 | XM_006718622 | ENSP00000265636 | 793 (aa) | XP_006718685 | Q5H9R7 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000110075 | Platelet Function Tests | 9.7357677E-006 | - |
| ENSG00000110075 | Platelet Function Tests | 1.0689400E-005 | - |
| ENSG00000110075 | Platelet Function Tests | 1.0007500E-005 | - |
| ENSG00000110075 | Child Development Disorders, Pervasive | 2.9029000E-005 | - |
| ENSG00000110075 | Bone Density | 1E-10 | 24945404 |
| ENSG00000110075 | Bone Density | 2E-10 | 24945404 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000110075 | rs7927592 | 11 | 68602492 | A | Total body bone mineral density | 29304378 | unit decrease | 0.0781 | EFO_0003923 |
| ENSG00000110075 | rs7927592 | 11 | 68602492 | A | Total body bone mineral density | 29304378 | unit decrease | 0.0791 | EFO_0003923 |
| ENSG00000110075 | rs12364620 | 11 | 68484655 | T | Total body bone mineral density (age 0-15) | 29304378 | unit increase | 0.0848 | EFO_0003923 |
| ENSG00000110075 | rs3758643 | 11 | 68599725 | T | Educational attainment (MTAG) | 30038396 | [0.0072-0.013] unit decrease | 0.0101 | EFO_0004784 |
| ENSG00000110075 | rs12284933 | 11 | 68552021 | A | TB-LM or TBLH-BMD (pleiotropy) | 28743860 | EFO_0003923|EFO_0004995 | ||
| ENSG00000110075 | rs6591341 | 11 | 68568010 | T | Lean body mass | 28743860 | [NR] unit decrease | 0.0741 | EFO_0004995 |
| ENSG00000110075 | rs58332211 | 11 | 68543404 | ? | Heel bone mineral density | 30048462 | [0.027-0.036] unit decrease | 0.0318014 | EFO_0009270 |
| ENSG00000110075 | rs73518807 | 11 | 68515518 | C | Educational attainment (years of education) | 30038396 | [0.0087-0.0169] unit decrease | 0.0128 | EFO_0004784 |
| ENSG00000110075 | rs2236708 | 11 | 68610577 | A | Educational attainment (years of education) | 30038396 | [0.0059-0.0121] unit decrease | 0.009 | EFO_0004784 |
| ENSG00000110075 | rs12283755 | 11 | 68604190 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000110075 | rs118074499 | 11 | 68533323 | ? | Hair color | 30595370 | EFO_0007822 | ||
| ENSG00000110075 | rs75356449 | 11 | 68574934 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000110075 | PPP6R3 | 100 | 82.051 | ENSG00000105063 | PPP6R1 | 100 | 64.331 | Homo_sapiens |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | - | IEA | Component |
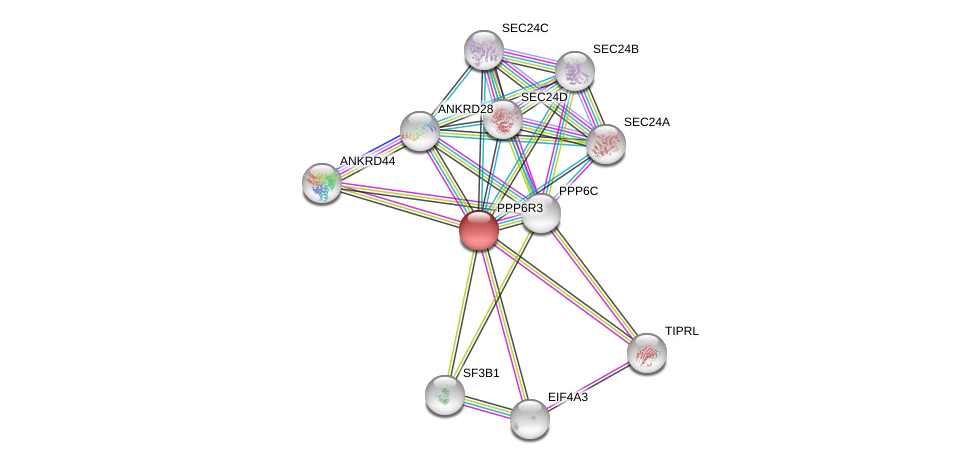
| GO:0005515 | protein binding | 18186651.21044950.21187329.24255178.28330616. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0019903 | protein phosphatase binding | 16716191. | IPI | Function |
| GO:0043666 | regulation of phosphoprotein phosphatase activity | 16716191. | IDA | Process |
| GO:0048208 | COPII vesicle coating | - | TAS | Process |