
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000547982 | LTA4H-205 | 986 | - | - | - (aa) | - | - |
| ENST00000228740 | LTA4H-201 | 2139 | XM_011538349 | ENSP00000228740 | 611 (aa) | XP_011536651 | P09960 |
| ENST00000537111 | LTA4H-203 | 1881 | - | - | - (aa) | - | - |
| ENST00000548375 | LTA4H-206 | 639 | - | - | - (aa) | - | - |
| ENST00000553041 | LTA4H-210 | 6109 | - | - | - (aa) | - | - |
| ENST00000552789 | LTA4H-209 | 2025 | - | ENSP00000449958 | 587 (aa) | - | P09960 |
| ENST00000548852 | LTA4H-207 | 1810 | - | ENSP00000449340 | 132 (aa) | - | B4DEH5 |
| ENST00000413268 | LTA4H-202 | 1908 | XM_005268871 | ENSP00000395051 | 508 (aa) | XP_005268928 | P09960 |
| ENST00000547393 | LTA4H-204 | 543 | - | - | - (aa) | - | - |
| ENST00000552091 | LTA4H-208 | 595 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000111144 | Varicose Veins | 2.49975621968999E-19 | - |
| ENSG00000111144 | Personality | 1.17e-005 | - |
| ENSG00000111144 | Platelet Function Tests | 5.9320979E-006 | - |
| ENSG00000111144 | Platelet Function Tests | 6.1858652E-006 | - |
| ENSG00000111144 | Platelet Function Tests | 9.3651473E-006 | - |
| ENSG00000111144 | Platelet Function Tests | 6.8013827E-006 | - |
| ENSG00000111144 | Platelet Function Tests | 6.9927571E-006 | - |
| ENSG00000111144 | Platelet Function Tests | 1.5141900E-005 | - |
| ENSG00000111144 | Platelet Function Tests | 2.5603000E-005 | - |
| ENSG00000111144 | Amino Acids | 5E-11 | 27005778 |
| ENSG00000111144 | Metabolism | 5E-11 | 27005778 |
| ENSG00000111144 | Proteins | 5E-11 | 27005778 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000111144 | rs2072510 | 12 | 96009421 | ? | Metabolite levels (small molecules and protein measures) | 27005778 | [0.048-0.089] unit decrease | 0.0689075 | EFO_0005664|EFO_0004747|EFO_0005134 |
| ENSG00000111144 | rs2247570 | 12 | 96028599 | T | Blood protein levels | 30072576 | [0.071-0.177] unit decrease | 0.124 | EFO_0007937 |
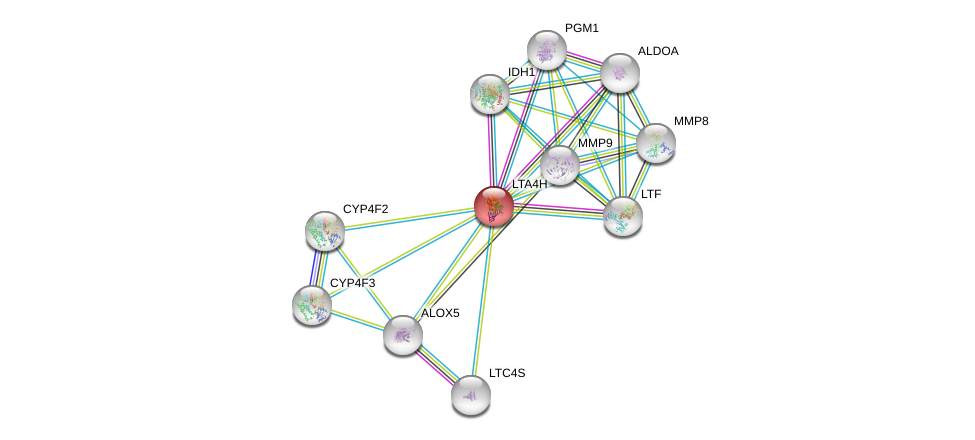
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0004177 | aminopeptidase activity | 9774412.15078870.18804029. | IDA | Function |
| GO:0004301 | epoxide hydrolase activity | 21873635. | IBA | Function |
| GO:0004301 | epoxide hydrolase activity | 15078870. | IDA | Function |
| GO:0004301 | epoxide hydrolase activity | 11675384. | IMP | Function |
| GO:0004301 | epoxide hydrolase activity | 11154734. | TAS | Function |
| GO:0004463 | leukotriene-A4 hydrolase activity | 21873635. | IBA | Function |
| GO:0004463 | leukotriene-A4 hydrolase activity | 9774412.15078870. | IDA | Function |
| GO:0005515 | protein binding | 21044950. | IPI | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006508 | proteolysis | - | IEA | Process |
| GO:0006691 | leukotriene metabolic process | - | TAS | Process |
| GO:0008233 | peptidase activity | 1881903. | TAS | Function |
| GO:0008270 | zinc ion binding | 15078870.18804029. | IDA | Function |
| GO:0008270 | zinc ion binding | 11675384. | IMP | Function |
| GO:0019370 | leukotriene biosynthetic process | 21873635. | IBA | Process |
| GO:0019370 | leukotriene biosynthetic process | 9774412.15078870. | IDA | Process |
| GO:0019370 | leukotriene biosynthetic process | 11675384. | IMP | Process |
| GO:0042759 | long-chain fatty acid biosynthetic process | - | TAS | Process |
| GO:0043171 | peptide catabolic process | 9774412.15078870.18804029. | IDA | Process |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0044255 | cellular lipid metabolic process | 21873635. | IBA | Process |
| GO:0044267 | cellular protein metabolic process | 11675384. | IMP | Process |
| GO:0070006 | metalloaminopeptidase activity | 11675384. | IMP | Function |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| GO:1904724 | tertiary granule lumen | - | TAS | Component |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |