
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000345345 | Exo_endo_phos | PF03372.23 | 3.7e-08 | 1 | 1 |
| ENSP00000367440 | Exo_endo_phos | PF03372.23 | 6.2e-08 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000478285 | TDP2-203 | 857 | - | - | - (aa) | - | - |
| ENST00000478507 | TDP2-204 | 573 | - | - | - (aa) | - | - |
| ENST00000341060 | TDP2-201 | 2120 | - | ENSP00000345345 | 304 (aa) | - | X6R5A3 |
| ENST00000480495 | TDP2-205 | 714 | - | - | - (aa) | - | - |
| ENST00000378198 | TDP2-202 | 2071 | - | ENSP00000367440 | 362 (aa) | - | O95551 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000111802 | rs3181245 | 6 | 24651092 | C | Colorectal or endometrial cancer | 26621817 | [1.07-1.16] | 1.12 | EFO_0005842|EFO_0004230 |
| ENSG00000111802 | rs3181238 | 6 | 24654215 | ? | Serum alkaline phosphatase levels | 29403010 | [0.024-0.041] unit increase novel | 0.03243 | EFO_0004533 |
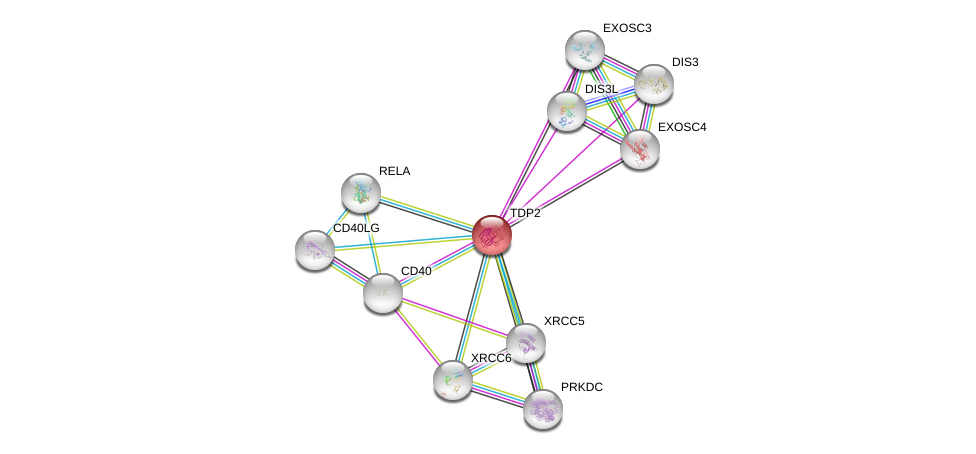
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000111802 | TDP2 | 86 | 53.208 | ENSAPOG00000018379 | - | 91 | 53.208 | Acanthochromis_polyacanthus |
| ENSG00000111802 | TDP2 | 92 | 86.738 | ENSAMEG00000010230 | TDP2 | 99 | 86.738 | Ailuropoda_melanoleuca |
| ENSG00000111802 | TDP2 | 60 | 52.717 | ENSAOCG00000010081 | - | 81 | 52.717 | Amphiprion_ocellaris |
| ENSG00000111802 | TDP2 | 95 | 49.488 | ENSAPEG00000004208 | - | 99 | 49.488 | Amphiprion_percula |
| ENSG00000111802 | TDP2 | 89 | 47.445 | ENSATEG00000010663 | - | 89 | 47.445 | Anabas_testudineus |
| ENSG00000111802 | TDP2 | 93 | 65.970 | ENSACAG00000002332 | TDP2 | 92 | 65.680 | Anolis_carolinensis |
| ENSG00000111802 | TDP2 | 100 | 91.713 | ENSANAG00000020033 | TDP2 | 100 | 91.713 | Aotus_nancymaae |
| ENSG00000111802 | TDP2 | 88 | 53.333 | ENSACLG00000016148 | tdp2b | 95 | 53.333 | Astatotilapia_calliptera |
| ENSG00000111802 | TDP2 | 92 | 48.582 | ENSACLG00000016132 | tdp2b | 95 | 48.582 | Astatotilapia_calliptera |
| ENSG00000111802 | TDP2 | 97 | 48.822 | ENSACLG00000016174 | tdp2b | 97 | 48.822 | Astatotilapia_calliptera |
| ENSG00000111802 | TDP2 | 91 | 56.679 | ENSAMXG00000011775 | tdp2b | 95 | 51.934 | Astyanax_mexicanus |
| ENSG00000111802 | TDP2 | 91 | 48.410 | ENSAMXG00000036549 | tdp2a | 92 | 48.410 | Astyanax_mexicanus |
| ENSG00000111802 | TDP2 | 94 | 85.714 | ENSBTAG00000000365 | TDP2 | 94 | 85.714 | Bos_taurus |
| ENSG00000111802 | TDP2 | 90 | 30.699 | WBGene00013405 | tdpt-1 | 85 | 30.699 | Caenorhabditis_elegans |
| ENSG00000111802 | TDP2 | 100 | 90.608 | ENSCJAG00000021963 | - | 100 | 90.608 | Callithrix_jacchus |
| ENSG00000111802 | TDP2 | 100 | 91.989 | ENSCJAG00000006074 | TDP2 | 100 | 91.989 | Callithrix_jacchus |
| ENSG00000111802 | TDP2 | 92 | 87.814 | ENSCAFG00000010428 | TDP2 | 99 | 85.294 | Canis_familiaris |
| ENSG00000111802 | TDP2 | 92 | 87.814 | ENSCAFG00020009994 | TDP2 | 96 | 87.814 | Canis_lupus_dingo |
| ENSG00000111802 | TDP2 | 92 | 87.900 | ENSCHIG00000015294 | - | 95 | 87.900 | Capra_hircus |
| ENSG00000111802 | TDP2 | 93 | 89.437 | ENSTSYG00000037199 | TDP2 | 100 | 89.644 | Carlito_syrichta |
| ENSG00000111802 | TDP2 | 100 | 85.635 | ENSCPOG00000006390 | TDP2 | 95 | 86.804 | Cavia_porcellus |
| ENSG00000111802 | TDP2 | 100 | 93.646 | ENSCCAG00000022734 | TDP2 | 100 | 93.646 | Cebus_capucinus |
| ENSG00000111802 | TDP2 | 100 | 95.724 | ENSCATG00000022076 | TDP2 | 100 | 95.724 | Cercocebus_atys |
| ENSG00000111802 | TDP2 | 100 | 83.978 | ENSCLAG00000002072 | TDP2 | 95 | 86.217 | Chinchilla_lanigera |
| ENSG00000111802 | TDP2 | 100 | 95.856 | ENSCSAG00000007769 | TDP2 | 100 | 95.856 | Chlorocebus_sabaeus |
| ENSG00000111802 | TDP2 | 95 | 69.096 | ENSCPBG00000006551 | TDP2 | 87 | 69.208 | Chrysemys_picta_bellii |
| ENSG00000111802 | TDP2 | 100 | 95.395 | ENSCANG00000040122 | TDP2 | 100 | 95.395 | Colobus_angolensis_palliatus |
| ENSG00000111802 | TDP2 | 91 | 73.741 | ENSCGRG00001000384 | Tdp2 | 92 | 69.208 | Cricetulus_griseus_chok1gshd |
| ENSG00000111802 | TDP2 | 91 | 73.741 | ENSCGRG00000010250 | Tdp2 | 94 | 67.155 | Cricetulus_griseus_crigri |
| ENSG00000111802 | TDP2 | 93 | 53.714 | ENSCVAG00000003114 | tdp2b | 95 | 53.143 | Cyprinodon_variegatus |
| ENSG00000111802 | TDP2 | 84 | 47.692 | ENSDARG00000069941 | tdp2a | 58 | 47.692 | Danio_rerio |
| ENSG00000111802 | TDP2 | 92 | 55.017 | ENSDARG00000035954 | tdp2b | 94 | 51.648 | Danio_rerio |
| ENSG00000111802 | TDP2 | 100 | 85.912 | ENSDNOG00000044082 | TDP2 | 93 | 88.270 | Dasypus_novemcinctus |
| ENSG00000111802 | TDP2 | 91 | 85.971 | ENSETEG00000019096 | TDP2 | 100 | 85.971 | Echinops_telfairi |
| ENSG00000111802 | TDP2 | 87 | 53.008 | ENSEBUG00000006535 | tdp2a | 62 | 53.008 | Eptatretus_burgeri |
| ENSG00000111802 | TDP2 | 94 | 87.390 | ENSEASG00005007656 | TDP2 | 94 | 87.390 | Equus_asinus_asinus |
| ENSG00000111802 | TDP2 | 94 | 87.390 | ENSECAG00000014264 | TDP2 | 94 | 87.390 | Equus_caballus |
| ENSG00000111802 | TDP2 | 100 | 80.387 | ENSEEUG00000010776 | TDP2 | 94 | 81.818 | Erinaceus_europaeus |
| ENSG00000111802 | TDP2 | 94 | 46.735 | ENSELUG00000009321 | tdp2a | 94 | 46.333 | Esox_lucius |
| ENSG00000111802 | TDP2 | 92 | 53.039 | ENSELUG00000016449 | tdp2b | 95 | 53.039 | Esox_lucius |
| ENSG00000111802 | TDP2 | 100 | 84.530 | ENSFCAG00000025612 | TDP2 | 94 | 86.217 | Felis_catus |
| ENSG00000111802 | TDP2 | 91 | 64.029 | ENSFALG00000004710 | TDP2 | 94 | 59.649 | Ficedula_albicollis |
| ENSG00000111802 | TDP2 | 96 | 85.616 | ENSFDAG00000006512 | TDP2 | 97 | 88.214 | Fukomys_damarensis |
| ENSG00000111802 | TDP2 | 81 | 51.613 | ENSFHEG00000013560 | - | 84 | 51.613 | Fundulus_heteroclitus |
| ENSG00000111802 | TDP2 | 91 | 69.784 | ENSGALG00000013637 | TDP2 | 92 | 64.620 | Gallus_gallus |
| ENSG00000111802 | TDP2 | 97 | 68.466 | ENSGAGG00000002853 | TDP2 | 87 | 69.501 | Gopherus_agassizii |
| ENSG00000111802 | TDP2 | 100 | 99.448 | ENSGGOG00000014237 | TDP2 | 100 | 99.448 | Gorilla_gorilla |
| ENSG00000111802 | TDP2 | 66 | 56.436 | ENSHBUG00000005421 | - | 83 | 57.711 | Haplochromis_burtoni |
| ENSG00000111802 | TDP2 | 91 | 56.475 | ENSHBUG00000002714 | tdp2b | 95 | 54.467 | Haplochromis_burtoni |
| ENSG00000111802 | TDP2 | 100 | 83.978 | ENSHGLG00000008655 | TDP2 | 95 | 84.751 | Heterocephalus_glaber_female |
| ENSG00000111802 | TDP2 | 100 | 83.978 | ENSHGLG00100019087 | TDP2 | 95 | 84.751 | Heterocephalus_glaber_male |
| ENSG00000111802 | TDP2 | 94 | 53.463 | ENSIPUG00000013691 | tdp2b | 94 | 53.824 | Ictalurus_punctatus |
| ENSG00000111802 | TDP2 | 98 | 64.326 | ENSJJAG00000021685 | Tdp2 | 97 | 66.272 | Jaculus_jaculus |
| ENSG00000111802 | TDP2 | 85 | 50.575 | ENSKMAG00000015485 | - | 84 | 50.575 | Kryptolebias_marmoratus |
| ENSG00000111802 | TDP2 | 91 | 63.669 | ENSLACG00000008852 | TDP2 | 95 | 58.857 | Latimeria_chalumnae |
| ENSG00000111802 | TDP2 | 91 | 58.333 | ENSLOCG00000005817 | tdp2b | 98 | 58.333 | Lepisosteus_oculatus |
| ENSG00000111802 | TDP2 | 94 | 87.097 | ENSLAFG00000002567 | TDP2 | 97 | 87.097 | Loxodonta_africana |
| ENSG00000111802 | TDP2 | 100 | 95.724 | ENSMFAG00000045296 | TDP2 | 100 | 95.724 | Macaca_fascicularis |
| ENSG00000111802 | TDP2 | 100 | 94.475 | ENSMMUG00000009803 | TDP2 | 100 | 94.475 | Macaca_mulatta |
| ENSG00000111802 | TDP2 | 100 | 95.395 | ENSMNEG00000038003 | TDP2 | 100 | 95.395 | Macaca_nemestrina |
| ENSG00000111802 | TDP2 | 100 | 96.382 | ENSMLEG00000028883 | TDP2 | 100 | 96.382 | Mandrillus_leucophaeus |
| ENSG00000111802 | TDP2 | 91 | 55.755 | ENSMAMG00000013654 | tdp2b | 94 | 54.386 | Mastacembelus_armatus |
| ENSG00000111802 | TDP2 | 93 | 53.890 | ENSMZEG00005003950 | tdp2b | 95 | 53.890 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 97 | 46.801 | ENSMZEG00005004103 | tdp2b | 97 | 46.801 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 87 | 53.008 | ENSMZEG00005004153 | tdp2b | 91 | 53.008 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 82 | 52.778 | ENSMZEG00005004055 | tdp2b | 83 | 52.778 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 83 | 47.244 | ENSMZEG00005003980 | - | 82 | 47.244 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 98 | 48.333 | ENSMZEG00005004126 | tdp2b | 96 | 48.333 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 90 | 51.636 | ENSMZEG00005004077 | tdp2b | 96 | 50.709 | Maylandia_zebra |
| ENSG00000111802 | TDP2 | 91 | 70.504 | ENSMGAG00000008014 | TDP2 | 91 | 65.689 | Meleagris_gallopavo |
| ENSG00000111802 | TDP2 | 91 | 73.022 | ENSMAUG00000014451 | Tdp2 | 92 | 68.328 | Mesocricetus_auratus |
| ENSG00000111802 | TDP2 | 100 | 88.705 | ENSMICG00000000039 | TDP2 | 100 | 87.789 | Microcebus_murinus |
| ENSG00000111802 | TDP2 | 91 | 74.820 | ENSMOCG00000002823 | Tdp2 | 92 | 70.381 | Microtus_ochrogaster |
| ENSG00000111802 | TDP2 | 89 | 65.152 | ENSMODG00000011653 | TDP2 | 95 | 67.203 | Monodelphis_domestica |
| ENSG00000111802 | TDP2 | 98 | 46.689 | ENSMALG00000005368 | - | 98 | 46.689 | Monopterus_albus |
| ENSG00000111802 | TDP2 | 91 | 74.101 | MGP_CAROLIEiJ_G0018354 | Tdp2 | 91 | 68.035 | Mus_caroli |
| ENSG00000111802 | TDP2 | 91 | 75.180 | ENSMUSG00000035958 | Tdp2 | 91 | 68.915 | Mus_musculus |
| ENSG00000111802 | TDP2 | 91 | 73.741 | MGP_PahariEiJ_G0019417 | Tdp2 | 91 | 66.862 | Mus_pahari |
| ENSG00000111802 | TDP2 | 91 | 74.820 | MGP_SPRETEiJ_G0019220 | Tdp2 | 91 | 68.622 | Mus_spretus |
| ENSG00000111802 | TDP2 | 91 | 88.129 | ENSMPUG00000004516 | TDP2 | 95 | 84.257 | Mustela_putorius_furo |
| ENSG00000111802 | TDP2 | 91 | 82.079 | ENSMLUG00000013506 | TDP2 | 94 | 77.353 | Myotis_lucifugus |
| ENSG00000111802 | TDP2 | 98 | 64.407 | ENSNGAG00000001375 | Tdp2 | 92 | 65.588 | Nannospalax_galili |
| ENSG00000111802 | TDP2 | 100 | 98.619 | ENSNLEG00000002447 | TDP2 | 100 | 98.619 | Nomascus_leucogenys |
| ENSG00000111802 | TDP2 | 96 | 83.862 | ENSOPRG00000016299 | TDP2 | 97 | 84.164 | Ochotona_princeps |
| ENSG00000111802 | TDP2 | 92 | 91.039 | ENSODEG00000014018 | TDP2 | 95 | 91.039 | Octodon_degus |
| ENSG00000111802 | TDP2 | 98 | 85.311 | ENSOCUG00000013773 | TDP2 | 94 | 86.510 | Oryctolagus_cuniculus |
| ENSG00000111802 | TDP2 | 87 | 48.315 | ENSORLG00000001661 | - | 97 | 48.315 | Oryzias_latipes |
| ENSG00000111802 | TDP2 | 87 | 48.315 | ENSORLG00020004634 | - | 97 | 48.315 | Oryzias_latipes_hni |
| ENSG00000111802 | TDP2 | 87 | 43.446 | ENSORLG00015008190 | - | 97 | 43.446 | Oryzias_latipes_hsok |
| ENSG00000111802 | TDP2 | 87 | 46.792 | ENSOMEG00000015899 | - | 92 | 49.597 | Oryzias_melastigma |
| ENSG00000111802 | TDP2 | 100 | 85.635 | ENSOGAG00000025303 | TDP2 | 93 | 87.390 | Otolemur_garnettii |
| ENSG00000111802 | TDP2 | 100 | 99.448 | ENSPPAG00000029023 | TDP2 | 100 | 99.448 | Pan_paniscus |
| ENSG00000111802 | TDP2 | 100 | 83.978 | ENSPPRG00000015398 | TDP2 | 94 | 85.630 | Panthera_pardus |
| ENSG00000111802 | TDP2 | 99 | 82.178 | ENSPTIG00000018574 | TDP2 | 95 | 87.050 | Panthera_tigris_altaica |
| ENSG00000111802 | TDP2 | 100 | 99.448 | ENSPTRG00000017778 | TDP2 | 100 | 99.448 | Pan_troglodytes |
| ENSG00000111802 | TDP2 | 100 | 95.066 | ENSPANG00000017590 | TDP2 | 100 | 95.066 | Papio_anubis |
| ENSG00000111802 | TDP2 | 92 | 53.659 | ENSPKIG00000006483 | tdp2b | 94 | 51.989 | Paramormyrops_kingsleyae |
| ENSG00000111802 | TDP2 | 92 | 71.326 | ENSPSIG00000004948 | TDP2 | 100 | 69.966 | Pelodiscus_sinensis |
| ENSG00000111802 | TDP2 | 91 | 74.820 | ENSPEMG00000001788 | Tdp2 | 91 | 70.088 | Peromyscus_maniculatus_bairdii |
| ENSG00000111802 | TDP2 | 81 | 47.148 | ENSPMAG00000001570 | tdp2b | 96 | 47.529 | Petromyzon_marinus |
| ENSG00000111802 | TDP2 | 91 | 72.662 | ENSPCIG00000007316 | TDP2 | 89 | 68.035 | Phascolarctos_cinereus |
| ENSG00000111802 | TDP2 | 81 | 50.400 | ENSPFOG00000017990 | - | 90 | 50.400 | Poecilia_formosa |
| ENSG00000111802 | TDP2 | 81 | 50.000 | ENSPLAG00000019439 | - | 79 | 50.000 | Poecilia_latipinna |
| ENSG00000111802 | TDP2 | 81 | 50.800 | ENSPMEG00000010824 | - | 94 | 46.528 | Poecilia_mexicana |
| ENSG00000111802 | TDP2 | 100 | 98.343 | ENSPPYG00000016282 | TDP2 | 92 | 98.343 | Pongo_abelii |
| ENSG00000111802 | TDP2 | 100 | 90.331 | ENSPCOG00000021415 | TDP2 | 100 | 90.331 | Propithecus_coquereli |
| ENSG00000111802 | TDP2 | 87 | 49.291 | ENSPNYG00000011504 | tdp2b | 90 | 49.291 | Pundamilia_nyererei |
| ENSG00000111802 | TDP2 | 69 | 49.282 | ENSPNYG00000001944 | - | 83 | 49.282 | Pundamilia_nyererei |
| ENSG00000111802 | TDP2 | 93 | 52.738 | ENSPNYG00000011532 | tdp2b | 95 | 52.738 | Pundamilia_nyererei |
| ENSG00000111802 | TDP2 | 86 | 50.385 | ENSPNAG00000022162 | tdp2a | 66 | 50.385 | Pygocentrus_nattereri |
| ENSG00000111802 | TDP2 | 94 | 71.777 | ENSRNOG00000018246 | Tdp2 | 92 | 68.328 | Rattus_norvegicus |
| ENSG00000111802 | TDP2 | 100 | 95.066 | ENSRBIG00000037119 | TDP2 | 100 | 95.066 | Rhinopithecus_bieti |
| ENSG00000111802 | TDP2 | 100 | 94.737 | ENSRROG00000005025 | TDP2 | 100 | 94.737 | Rhinopithecus_roxellana |
| ENSG00000111802 | TDP2 | 100 | 91.436 | ENSSBOG00000028734 | TDP2 | 100 | 90.789 | Saimiri_boliviensis_boliviensis |
| ENSG00000111802 | TDP2 | 99 | 65.489 | ENSSHAG00000002506 | TDP2 | 91 | 67.544 | Sarcophilus_harrisii |
| ENSG00000111802 | TDP2 | 84 | 52.756 | ENSSFOG00015011322 | - | 71 | 53.175 | Scleropages_formosus |
| ENSG00000111802 | TDP2 | 91 | 79.211 | ENSSARG00000013547 | TDP2 | 100 | 79.211 | Sorex_araneus |
| ENSG00000111802 | TDP2 | 94 | 64.857 | ENSSPUG00000008288 | TDP2 | 95 | 64.857 | Sphenodon_punctatus |
| ENSG00000111802 | TDP2 | 100 | 86.188 | ENSSSCG00000001092 | TDP2 | 94 | 87.683 | Sus_scrofa |
| ENSG00000111802 | TDP2 | 86 | 68.199 | ENSTGUG00000009035 | TDP2 | 100 | 68.199 | Taeniopygia_guttata |
| ENSG00000111802 | TDP2 | 99 | 48.185 | ENSTRUG00000012218 | - | 99 | 48.185 | Takifugu_rubripes |
| ENSG00000111802 | TDP2 | 93 | 53.561 | ENSTRUG00000014347 | tdp2b | 96 | 53.258 | Takifugu_rubripes |
| ENSG00000111802 | TDP2 | 96 | 52.066 | ENSTNIG00000017792 | tdp2b | 96 | 52.676 | Tetraodon_nigroviridis |
| ENSG00000111802 | TDP2 | 83 | 54.023 | ENSTNIG00000010260 | TDP2 | 99 | 54.023 | Tetraodon_nigroviridis |
| ENSG00000111802 | TDP2 | 100 | 84.530 | ENSTTRG00000016376 | TDP2 | 94 | 86.510 | Tursiops_truncatus |
| ENSG00000111802 | TDP2 | 99 | 82.392 | ENSUAMG00000025939 | TDP2 | 95 | 80.352 | Ursus_americanus |
| ENSG00000111802 | TDP2 | 100 | 80.921 | ENSUMAG00000020111 | TDP2 | 100 | 80.921 | Ursus_maritimus |
| ENSG00000111802 | TDP2 | 98 | 61.141 | ENSXETG00000014826 | tdp2 | 95 | 62.147 | Xenopus_tropicalis |
| ENSG00000111802 | TDP2 | 81 | 50.996 | ENSXCOG00000010778 | - | 69 | 50.996 | Xiphophorus_couchianus |
| ENSG00000111802 | TDP2 | 81 | 50.996 | ENSXMAG00000022252 | - | 72 | 50.996 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | 22405347.22822062. | IDA | Function |
| GO:0000287 | magnesium ion binding | 19794497. | TAS | Function |
| GO:0003697 | single-stranded DNA binding | 21873635. | IBA | Function |
| GO:0003697 | single-stranded DNA binding | 22822062. | IDA | Function |
| GO:0003714 | transcription corepressor activity | 10764746. | TAS | Function |
| GO:0004518 | nuclease activity | - | IEA | Function |
| GO:0005515 | protein binding | 18039968. | IPI | Function |
| GO:0005634 | nucleus | 22908287. | IDA | Component |
| GO:0005654 | nucleoplasm | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 22908287. | IDA | Component |
| GO:0006302 | double-strand break repair | 21873635. | IBA | Process |
| GO:0006302 | double-strand break repair | 19794497. | IDA | Process |
| GO:0006302 | double-strand break repair | 21030584. | IMP | Process |
| GO:0007166 | cell surface receptor signaling pathway | 10764746. | TAS | Process |
| GO:0016235 | aggresome | - | IDA | Component |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0016605 | PML body | 21873635. | IBA | Component |
| GO:0016605 | PML body | 19794497. | IDA | Component |
| GO:0030145 | manganese ion binding | 22822062. | IDA | Function |
| GO:0036317 | tyrosyl-RNA phosphodiesterase activity | 22908287. | IDA | Function |
| GO:0048666 | neuron development | 24658003. | IMP | Process |
| GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity | 21873635. | IBA | Function |
| GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity | 19794497.22405347.22822062. | IDA | Function |
| GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity | 21030584. | IMP | Function |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | - | IEA | Process |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| SARC |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| ESCA | ||
| PAAD | ||
| BLCA | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| OV |