
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000649854 | MOGS-219 | 2304 | - | ENSP00000496797 | 715 (aa) | - | - |
| ENST00000462189 | MOGS-206 | 2400 | - | - | - (aa) | - | - |
| ENST00000452063 | MOGS-205 | 2433 | - | ENSP00000388201 | 731 (aa) | - | Q13724 |
| ENST00000648810 | MOGS-215 | 2126 | - | ENSP00000496949 | 562 (aa) | - | UPI000048BAEA |
| ENST00000647915 | MOGS-213 | 2269 | - | ENSP00000498123 | 53 (aa) | - | - |
| ENST00000647753 | MOGS-211 | 2445 | - | ENSP00000497318 | 159 (aa) | - | B4E3B8 |
| ENST00000489655 | MOGS-209 | 489 | - | - | - (aa) | - | - |
| ENST00000233616 | MOGS-201 | 2895 | - | ENSP00000233616 | 837 (aa) | - | Q13724 |
| ENST00000448666 | MOGS-204 | 2867 | - | ENSP00000410992 | 837 (aa) | - | C9J8D4 |
| ENST00000649777 | MOGS-218 | 2775 | - | - | - (aa) | - | - |
| ENST00000462443 | MOGS-207 | 2205 | - | ENSP00000497265 | 562 (aa) | - | UPI000048BAEA |
| ENST00000486036 | MOGS-208 | 811 | - | - | - (aa) | - | - |
| ENST00000647723 | MOGS-210 | 2606 | - | ENSP00000497924 | 208 (aa) | - | - |
| ENST00000648768 | MOGS-214 | 2862 | - | - | - (aa) | - | - |
| ENST00000649075 | MOGS-216 | 2649 | - | ENSP00000497836 | 294 (aa) | - | - |
| ENST00000647771 | MOGS-212 | 2469 | - | ENSP00000496788 | 75 (aa) | - | - |
| ENST00000414701 | MOGS-203 | 520 | - | ENSP00000396298 | 145 (aa) | - | C9JDQ1 |
| ENST00000649601 | MOGS-217 | 2316 | - | ENSP00000496796 | 113 (aa) | - | - |
| ENST00000650523 | MOGS-220 | 1332 | - | ENSP00000497143 | 428 (aa) | - | - |
| ENST00000409065 | MOGS-202 | 2668 | - | ENSP00000386493 | 209 (aa) | - | B8ZZE2 |
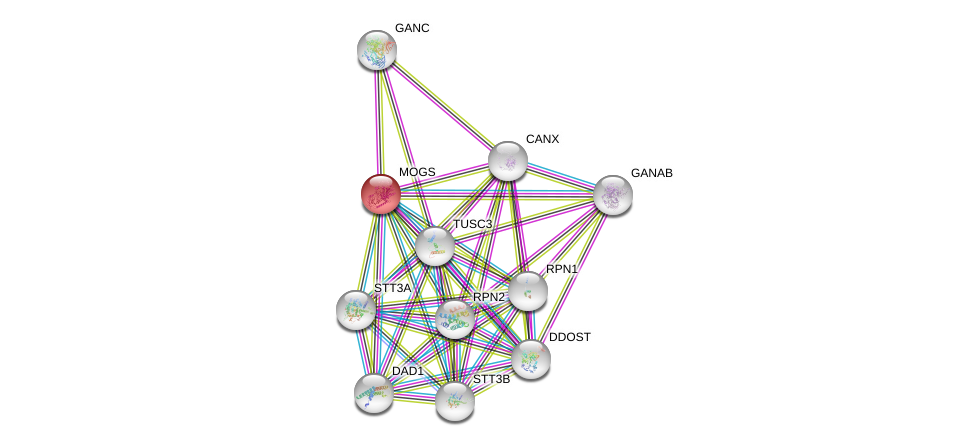
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000115275 | MOGS | 100 | 89.726 | ENSMUSG00000030036 | Mogs | 100 | 86.277 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004573 | mannosyl-oligosaccharide glucosidase activity | 21873635. | IBA | Function |
| GO:0004573 | mannosyl-oligosaccharide glucosidase activity | - | TAS | Function |
| GO:0005783 | endoplasmic reticulum | - | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | 21873635. | IBA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0006457 | protein folding | - | TAS | Process |
| GO:0006487 | protein N-linked glycosylation | 21873635. | IBA | Process |
| GO:0009311 | oligosaccharide metabolic process | - | IEA | Process |
| GO:0015926 | glucosidase activity | 7635146. | TAS | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0070062 | extracellular exosome | 19056867.19199708. | HDA | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| PCPG | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| OV |