
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000377488 | PARK7-202 | 783 | - | ENSP00000366708 | 189 (aa) | - | Q99497 |
| ENST00000497113 | PARK7-210 | 670 | - | - | - (aa) | - | - |
| ENST00000469225 | PARK7-207 | 711 | - | ENSP00000466756 | 160 (aa) | - | K7EN27 |
| ENST00000493373 | PARK7-208 | 624 | - | ENSP00000465404 | 189 (aa) | - | Q99497 |
| ENST00000493678 | PARK7-209 | 1088 | - | ENSP00000418770 | 189 (aa) | - | Q99497 |
| ENST00000338639 | PARK7-201 | 949 | - | ENSP00000340278 | 189 (aa) | - | Q99497 |
| ENST00000377493 | PARK7-204 | 795 | - | ENSP00000466242 | 169 (aa) | - | K7ELW0 |
| ENST00000460192 | PARK7-205 | 567 | - | - | - (aa) | - | - |
| ENST00000377491 | PARK7-203 | 977 | XM_005263424 | ENSP00000366711 | 189 (aa) | XP_005263481 | Q99497 |
| ENST00000465354 | PARK7-206 | 949 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05012 | Parkinson disease | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000116288 | Colitis, Ulcerative | 6E-14 | 26192919 |
| ENSG00000116288 | Colitis, Ulcerative | 5E-9 | 21297633 |
| ENSG00000116288 | Crohn Disease | 5E-9 | 26192919 |
| ENSG00000116288 | Inflammatory Bowel Diseases | 1E-12 | 28067908 |
| ENSG00000116288 | Stroke | 2E-7 | 24262325 |
| ENSG00000116288 | Colitis, Ulcerative | 4E-9 | 28067908 |
| ENSG00000116288 | Inflammatory Bowel Diseases | 1E-15 | 23128233 |
| ENSG00000116288 | Celiac Disease | 9E-8 | 20190752 |
| ENSG00000116288 | Crohn Disease | 3E-6 | 28067908 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000116288 | rs161802 | 1 | 7982766 | ? | Ischemic stroke | 24262325 | [1.08-1.16] | 1.1111 | EFO_0000712 |
| ENSG00000116288 | rs3766606 | 1 | 7962137 | C | Ulcerative colitis | 26192919 | 1.143554 | EFO_0000729 | |
| ENSG00000116288 | rs3766606 | 1 | 7962137 | A | Crohn's disease | 26192919 | 1.1060587 | EFO_0000384 | |
| ENSG00000116288 | rs17523802 | 1 | 7961680 | ? | Tonsillectomy | 28928442 | [0.033-0.076] unit increase | 0.0545 | EFO_0007924 |
| ENSG00000116288 | rs3766606 | 1 | 7962137 | ? | Crohn's disease | 28067908 | EFO_0000384 | ||
| ENSG00000116288 | rs3766606 | 1 | 7962137 | ? | Ulcerative colitis | 28067908 | EFO_0000729 | ||
| ENSG00000116288 | rs3766606 | 1 | 7962137 | ? | Inflammatory bowel disease | 28067908 | EFO_0003767 | ||
| ENSG00000116288 | rs35675666 | 1 | 7961913 | G | Ulcerative colitis | 21297633 | [1.02-1.15] | 1.08 | EFO_0000729 |
| ENSG00000116288 | rs35675666 | 1 | 7961913 | G | Inflammatory bowel disease | 23128233 | [1.07-1.156] | 1.112 | EFO_0003767 |
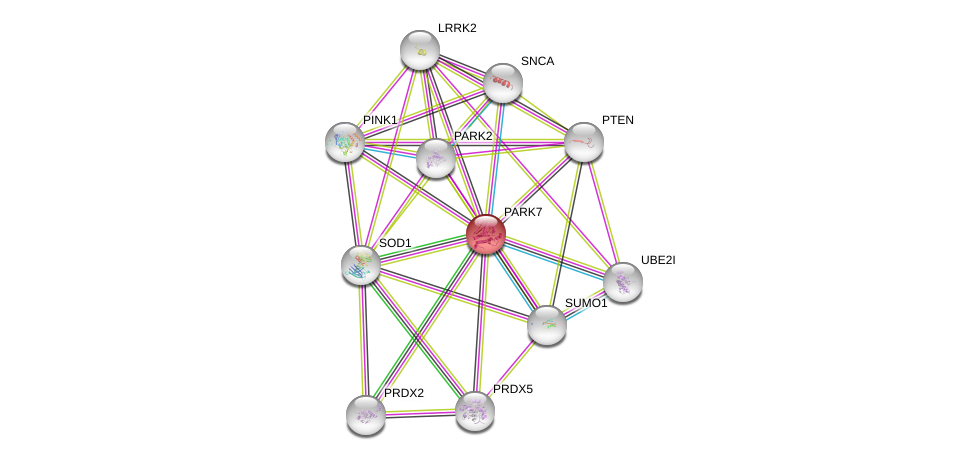
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000116288 | PARK7 | 100 | 91.534 | ENSMUSG00000028964 | Park7 | 100 | 95.238 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000785 | chromatin | 16731528. | IDA | Component |
| GO:0001047 | core promoter binding | 15790595. | IC | Function |
| GO:0001933 | negative regulation of protein phosphorylation | 15983381. | IGI | Process |
| GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | - | ISS | Process |
| GO:0003690 | double-stranded DNA binding | 22683601. | IDA | Function |
| GO:0003697 | single-stranded DNA binding | 22683601. | IDA | Function |
| GO:0003713 | transcription coactivator activity | 15790595. | IGI | Function |
| GO:0003713 | transcription coactivator activity | 16731528. | TAS | Function |
| GO:0003729 | mRNA binding | 18626009. | IDA | Function |
| GO:0005102 | signaling receptor binding | 21785459. | IPI | Function |
| GO:0005507 | copper ion binding | 23792957. | IDA | Function |
| GO:0005515 | protein binding | 11477070.15790595.15983381.16632486.17510388.19229105.19822128.20127688.21097510.21785459.22683601.23743200.24899725.24947010.25416956. | IPI | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 11477070.12446870.14662519.15790595.15983381.18711745.19822128.22683601. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 12446870.15983381.17510388.18711745.19822128. | IDA | Component |
| GO:0005739 | mitochondrion | 15790595.15944198.18711745.19229105.19822128.23743200. | IDA | Component |
| GO:0005739 | mitochondrion | 21785459. | IMP | Component |
| GO:0005747 | mitochondrial respiratory chain complex I | 19822128. | IDA | Component |
| GO:0005758 | mitochondrial intermembrane space | - | IEA | Component |
| GO:0005759 | mitochondrial matrix | - | IEA | Component |
| GO:0005783 | endoplasmic reticulum | - | IEA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 14662519.15944198.19229105.22683601. | IDA | Component |
| GO:0005829 | cytosol | 21785459. | IMP | Component |
| GO:0005886 | plasma membrane | - | IEA | Component |
| GO:0005913 | cell-cell adherens junction | 25468996. | HDA | Component |
| GO:0006281 | DNA repair | 28596309. | IDA | Process |
| GO:0006469 | negative regulation of protein kinase activity | 15983381. | IGI | Process |
| GO:0006508 | proteolysis | - | IEA | Process |
| GO:0006517 | protein deglycosylation | 25416785. | IDA | Process |
| GO:0006517 | protein deglycosylation | 27903648. | IDA | Process |
| GO:0006517 | protein deglycosylation | 26995087. | IMP | Process |
| GO:0006914 | autophagy | - | IEA | Process |
| GO:0006954 | inflammatory response | - | IEA | Process |
| GO:0007005 | mitochondrion organization | - | ISS | Process |
| GO:0007265 | Ras protein signal transduction | 14662519. | TAS | Process |
| GO:0007338 | single fertilization | - | IEA | Process |
| GO:0008134 | transcription factor binding | 21097510. | IPI | Function |
| GO:0008233 | peptidase activity | 20304780. | IDA | Function |
| GO:0008344 | adult locomotory behavior | - | IEA | Process |
| GO:0009438 | methylglyoxal metabolic process | 22523093. | IDA | Process |
| GO:0009438 | methylglyoxal metabolic process | 27903648. | IDA | Process |
| GO:0010273 | detoxification of copper ion | 23792957. | IMP | Process |
| GO:0010628 | positive regulation of gene expression | 24252804. | TAS | Process |
| GO:0010629 | negative regulation of gene expression | 22683601. | IDA | Process |
| GO:0016532 | superoxide dismutase copper chaperone activity | 24567322. | IDA | Function |
| GO:0016605 | PML body | 22683601. | IDA | Component |
| GO:0016684 | oxidoreductase activity, acting on peroxide as acceptor | 24567322. | IDA | Function |
| GO:0018323 | enzyme active site formation via L-cysteine sulfinic acid | - | IEA | Process |
| GO:0019249 | lactate biosynthetic process | 22523093. | IDA | Process |
| GO:0019899 | enzyme binding | 19703902.19822128.23743200. | IPI | Function |
| GO:0019900 | kinase binding | 19229105. | IPI | Function |
| GO:0019955 | cytokine binding | 21097510. | IPI | Function |
| GO:0030073 | insulin secretion | - | ISS | Process |
| GO:0030424 | axon | - | ISS | Component |
| GO:0031397 | negative regulation of protein ubiquitination | 17015834.24899725. | IDA | Process |
| GO:0032091 | negative regulation of protein binding | 11477070.16731528.17015834.24899725. | IDA | Process |
| GO:0032091 | negative regulation of protein binding | 15983381. | IGI | Process |
| GO:0032091 | negative regulation of protein binding | 21785459. | IMP | Process |
| GO:0032148 | activation of protein kinase B activity | 22492997. | IC | Process |
| GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 17015834. | IDA | Process |
| GO:0032757 | positive regulation of interleukin-8 production | 21097510. | IDA | Process |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 22492997. | IMP | Process |
| GO:0033234 | negative regulation of protein sumoylation | 16731528. | IDA | Process |
| GO:0033864 | positive regulation of NAD(P)H oxidase activity | - | ISS | Process |
| GO:0034599 | cellular response to oxidative stress | 15983381.19703902.20969476.22683601. | IDA | Process |
| GO:0034599 | cellular response to oxidative stress | 24899725. | IMP | Process |
| GO:0036470 | tyrosine 3-monooxygenase activator activity | 19703902. | IDA | Function |
| GO:0036471 | cellular response to glyoxal | 22523093. | IDA | Process |
| GO:0036478 | L-dopa decarboxylase activator activity | 19703902. | IDA | Function |
| GO:0036524 | protein deglycase activity | 21873635. | IBA | Function |
| GO:0036524 | protein deglycase activity | 25416785. | IDA | Function |
| GO:0036526 | peptidyl-cysteine deglycation | 25416785. | IDA | Process |
| GO:0036527 | peptidyl-arginine deglycation | 25416785. | IDA | Process |
| GO:0036528 | peptidyl-lysine deglycation | 25416785. | IDA | Process |
| GO:0036529 | protein deglycation, glyoxal removal | 21873635. | IBA | Process |
| GO:0036529 | protein deglycation, glyoxal removal | 25416785. | IDA | Process |
| GO:0036530 | protein deglycation, methylglyoxal removal | 25416785. | IDA | Process |
| GO:0036530 | protein deglycation, methylglyoxal removal | 27903648. | IDA | Process |
| GO:0036531 | glutathione deglycation | 25416785. | IDA | Process |
| GO:0042593 | glucose homeostasis | - | ISS | Process |
| GO:0042743 | hydrogen peroxide metabolic process | 20969476.24567322. | IDA | Process |
| GO:0042802 | identical protein binding | 15502874.15983381.24947010. | IPI | Function |
| GO:0042803 | protein homodimerization activity | 18711745.24144264.24567322. | IDA | Function |
| GO:0043066 | negative regulation of apoptotic process | 22523093. | IDA | Process |
| GO:0043523 | regulation of neuron apoptotic process | 18711745.20304780. | IDA | Process |
| GO:0043524 | negative regulation of neuron apoptotic process | 22511790. | IDA | Process |
| GO:0044297 | cell body | - | IEA | Component |
| GO:0044388 | small protein activating enzyme binding | 15983381. | IPI | Function |
| GO:0044390 | ubiquitin-like protein conjugating enzyme binding | 15983381. | IPI | Function |
| GO:0045121 | membrane raft | - | IEA | Component |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0045340 | mercury ion binding | 23792957. | IDA | Function |
| GO:0045560 | regulation of TRAIL receptor biosynthetic process | 21785459. | IMP | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21097510. | IDA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 15790595.17015834. | IGI | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 16731528.19703902.22492997. | IMP | Process |
| GO:0046295 | glycolate biosynthetic process | 22523093. | IDA | Process |
| GO:0046826 | negative regulation of protein export from nucleus | 15983381. | IGI | Process |
| GO:0048471 | perinuclear region of cytoplasm | 25468996. | IDA | Component |
| GO:0050681 | androgen receptor binding | 17510388. | IPI | Function |
| GO:0050727 | regulation of inflammatory response | - | ISS | Process |
| GO:0050787 | detoxification of mercury ion | 23792957. | IMP | Process |
| GO:0050821 | protein stabilization | 24947010. | IDA | Process |
| GO:0050821 | protein stabilization | 17015834.19229105. | IMP | Process |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | 17015834. | IMP | Process |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | 22492997. | TAS | Process |
| GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 24899725. | IDA | Process |
| GO:0051583 | dopamine uptake involved in synaptic transmission | - | IEA | Process |
| GO:0051881 | regulation of mitochondrial membrane potential | 23743200. | IMP | Process |
| GO:0051897 | positive regulation of protein kinase B signaling | 22492997. | IC | Process |
| GO:0051899 | membrane depolarization | - | IEA | Process |
| GO:0051920 | peroxiredoxin activity | - | IEA | Function |
| GO:0060081 | membrane hyperpolarization | - | IEA | Process |
| GO:0060548 | negative regulation of cell death | 14749723. | IDA | Process |
| GO:0060765 | regulation of androgen receptor signaling pathway | 11477070. | IDA | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:0070301 | cellular response to hydrogen peroxide | 14749723. | IDA | Process |
| GO:0070491 | repressing transcription factor binding | 15790595.16731528. | IPI | Function |
| GO:0090073 | positive regulation of protein homodimerization activity | 24947010. | IDA | Process |
| GO:0097110 | scaffold protein binding | 21785459. | IPI | Function |
| GO:0098793 | presynapse | - | IEA | Component |
| GO:0098869 | cellular oxidant detoxification | - | IEA | Process |
| GO:0106044 | guanine deglycation | 28596309. | IDA | Process |
| GO:0106045 | guanine deglycation, methylglyoxal removal | 28596309. | IDA | Process |
| GO:0106046 | guanine deglycation, glyoxal removal | 21873635. | IBA | Process |
| GO:0106046 | guanine deglycation, glyoxal removal | 28596309. | IDA | Process |
| GO:1900182 | positive regulation of protein localization to nucleus | 21097510. | IDA | Process |
| GO:1900182 | positive regulation of protein localization to nucleus | 22492997. | IMP | Process |
| GO:1901215 | negative regulation of neuron death | 22683601. | IDA | Process |
| GO:1901671 | positive regulation of superoxide dismutase activity | 24567322. | IDA | Process |
| GO:1901984 | negative regulation of protein acetylation | 22683601. | IDA | Process |
| GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | - | IEA | Process |
| GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 14652021. | IGI | Process |
| GO:1902903 | regulation of supramolecular fiber organization | 23626584. | TAS | Process |
| GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 19822128. | IMP | Process |
| GO:1903073 | negative regulation of death-inducing signaling complex assembly | 21785459. | IC | Process |
| GO:1903094 | negative regulation of protein K48-linked deubiquitination | 21097510. | IDA | Process |
| GO:1903122 | negative regulation of TRAIL-activated apoptotic signaling pathway | 21785459. | IMP | Process |
| GO:1903135 | cupric ion binding | 24567322. | IDA | Function |
| GO:1903136 | cuprous ion binding | 24144264.24567322. | IDA | Function |
| GO:1903168 | positive regulation of pyrroline-5-carboxylate reductase activity | 23743200. | IDA | Process |
| GO:1903178 | positive regulation of tyrosine 3-monooxygenase activity | 19703902. | IDA | Process |
| GO:1903181 | positive regulation of dopamine biosynthetic process | 16731528. | IC | Process |
| GO:1903181 | positive regulation of dopamine biosynthetic process | 19703902. | IDA | Process |
| GO:1903189 | glyoxal metabolic process | 22523093. | IDA | Process |
| GO:1903197 | positive regulation of L-dopa biosynthetic process | 16731528. | IMP | Process |
| GO:1903200 | positive regulation of L-dopa decarboxylase activity | 19703902. | IDA | Process |
| GO:1903202 | negative regulation of oxidative stress-induced cell death | 16632486. | IDA | Process |
| GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 22492997.23743200. | IMP | Process |
| GO:1903208 | negative regulation of hydrogen peroxide-induced neuron death | 15983381.24947010. | IDA | Process |
| GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 15790595. | IDA | Process |
| GO:1903384 | negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 14652021. | IGI | Process |
| GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | - | ISS | Process |
| GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | - | IEA | Process |
| GO:1903599 | positive regulation of autophagy of mitochondrion | 24531622. | NAS | Process |
| GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 14752510. | IDA | Process |
| GO:1990381 | ubiquitin-specific protease binding | 21097510. | IPI | Function |
| GO:2000157 | negative regulation of ubiquitin-specific protease activity | 21097510. | IDA | Process |
| GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity | - | IEA | Process |
| GO:2000679 | positive regulation of transcription regulatory region DNA binding | 22492997. | IMP | Process |
| GO:2000825 | positive regulation of androgen receptor activity | 17510388. | IMP | Process |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 21785459. | IMP | Process |
| GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 21785459. | IMP | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LGG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| CHOL | |||
| DLBC | |||
| GBM | |||
| HNSC | |||
| KIRC | |||
| KIRP | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| READ | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| ACC | ||
| ESCA | ||
| KIRP | ||
| PCPG | ||
| LAML | ||
| KICH | ||
| UVM |