
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000367466 | PLA2G4A-201 | 2875 | XM_005245267 | ENSP00000356436 | 749 (aa) | XP_005245324 | P47712 |
| ENST00000466600 | PLA2G4A-202 | 850 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00564 | Glycerophospholipid metabolism | KEGG |
| hsa00565 | Ether lipid metabolism | KEGG |
| hsa00590 | Arachidonic acid metabolism | KEGG |
| hsa00591 | Linoleic acid metabolism | KEGG |
| hsa00592 | alpha-Linolenic acid metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa04010 | MAPK signaling pathway | KEGG |
| hsa04014 | Ras signaling pathway | KEGG |
| hsa04072 | Phospholipase D signaling pathway | KEGG |
| hsa04217 | Necroptosis | KEGG |
| hsa04270 | Vascular smooth muscle contraction | KEGG |
| hsa04370 | VEGF signaling pathway | KEGG |
| hsa04611 | Platelet activation | KEGG |
| hsa04664 | Fc epsilon RI signaling pathway | KEGG |
| hsa04666 | Fc gamma R-mediated phagocytosis | KEGG |
| hsa04724 | Glutamatergic synapse | KEGG |
| hsa04726 | Serotonergic synapse | KEGG |
| hsa04730 | Long-term depression | KEGG |
| hsa04750 | Inflammatory mediator regulation of TRP channels | KEGG |
| hsa04912 | GnRH signaling pathway | KEGG |
| hsa04913 | Ovarian steroidogenesis | KEGG |
| hsa04921 | Oxytocin signaling pathway | KEGG |
| hsa05231 | Choline metabolism in cancer | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000116711 | Cholesterol, LDL | 3.27953964541687E-7 | 17903299 |
| ENSG00000116711 | Muscle, Skeletal | 9.9090000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 5.2920000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 5.5770000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 3.0900000E-007 | - |
| ENSG00000116711 | Muscle, Skeletal | 1.3720000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 1.1420000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 6.4220000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 3.0200000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 2.6250000E-007 | - |
| ENSG00000116711 | Muscle, Skeletal | 2.8230000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 1.9410000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 4.0260000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 3.2840000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 6.8900000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 7.2330000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 1.1200000E-006 | - |
| ENSG00000116711 | Muscle, Skeletal | 7.8960000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 6.5720000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 1.0890000E-006 | - |
| ENSG00000116711 | Muscle, Skeletal | 5.6940000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 7.5600000E-005 | - |
| ENSG00000116711 | Muscle, Skeletal | 8.2600000E-005 | - |
| ENSG00000116711 | Lipoprotein(a) | 9.7740000E-005 | - |
| ENSG00000116711 | Lipoprotein(a) | 5.4440000E-007 | - |
| ENSG00000116711 | Urine | 3.9483822E-006 | - |
| ENSG00000116711 | Blood Pressure | 8.2994520E-005 | - |
| ENSG00000116711 | Blood Pressure | 3.0252510E-006 | - |
| ENSG00000116711 | Body Mass Index | 9.1584512E-005 | - |
| ENSG00000116711 | Body Mass Index | 8.4753884E-005 | - |
| ENSG00000116711 | Platelet Function Tests | 1.8161500E-005 | - |
| ENSG00000116711 | Platelet Function Tests | 2.8354695E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 2.7777360E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 2.3508931E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 6.0759671E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 5.9546754E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 5.1498934E-006 | - |
| ENSG00000116711 | Platelet Function Tests | 2.2204460E-016 | - |
| ENSG00000116711 | Platelet Function Tests | 2.2204460E-016 | - |
| ENSG00000116711 | Child Development Disorders, Pervasive | 1.0935600E-005 | - |
| ENSG00000116711 | Child Development Disorders, Pervasive | 1.4833000E-005 | - |
| ENSG00000116711 | Autistic Disorder | 6.2900000E-006 | - |
| ENSG00000116711 | Child Development Disorders, Pervasive | 1.1864000E-005 | - |
| ENSG00000116711 | Child Development Disorders, Pervasive | 1.0050000E-005 | - |
| ENSG00000116711 | Child Development Disorders, Pervasive | 1.8346600E-005 | - |
| ENSG00000116711 | Interleukin-6 | 8.6980000E-006 | - |
| ENSG00000116711 | Interleukin-6 | 3.9120000E-005 | - |
| ENSG00000116711 | Triiodothyronine | 2.0540000E-005 | - |
| ENSG00000116711 | Iron | 1.0470000E-005 | - |
| ENSG00000116711 | Vitamin A | 3.7360000E-006 | - |
| ENSG00000116711 | Schizophrenia | 7E-6 | 26198764 |
| ENSG00000116711 | Hydroxyindoleacetic Acid | 7E-6 | 23319000 |
| ENSG00000116711 | Crohn Disease | 4E-9 | 26192919 |
| ENSG00000116711 | Inflammatory Bowel Diseases | 2E-7 | 26192919 |
| ENSG00000116711 | Epilepsy | 9E-6 | 22949513 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000116711 | rs114333818 | 1 | 186856584 | A | Schizophrenia | 26198764 | [NR] | 1.3513513 | EFO_0000692 |
| ENSG00000116711 | rs10798069 | 1 | 186906327 | A | Crohn's disease | 26192919 | 1.0729233 | EFO_0000384 | |
| ENSG00000116711 | rs10798069 | 1 | 186906327 | C | Inflammatory bowel disease | 26192919 | 1.0524286 | EFO_0003767 | |
| ENSG00000116711 | rs12720541 | 1 | 186900940 | T | Generalized epilepsy | 22949513 | [1.10-1.35] | 1.21 | EFO_0000474 |
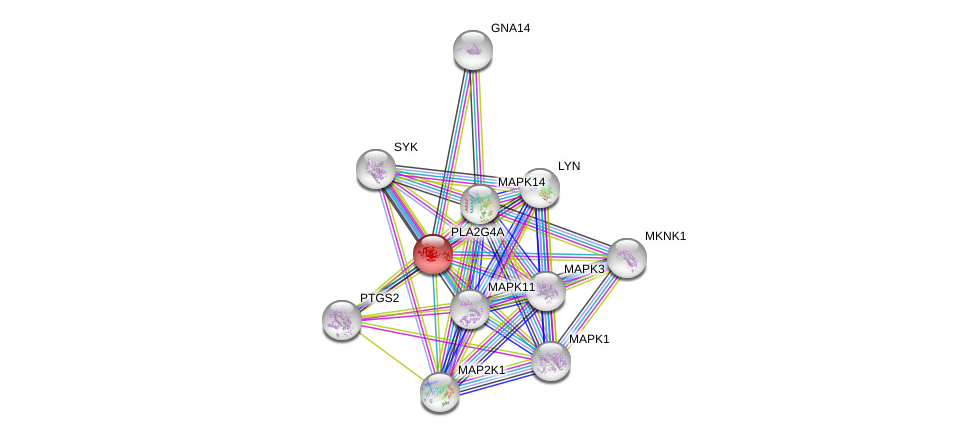
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004622 | lysophospholipase activity | - | IEA | Function |
| GO:0004623 | phospholipase A2 activity | 21873635. | IBA | Function |
| GO:0004623 | phospholipase A2 activity | 8702602. | IDA | Function |
| GO:0004623 | phospholipase A2 activity | 8381049. | NAS | Function |
| GO:0005509 | calcium ion binding | 21873635. | IBA | Function |
| GO:0005509 | calcium ion binding | 9430701.9665851. | IDA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 21873635. | IBA | Function |
| GO:0005544 | calcium-dependent phospholipid binding | 9665851. | IDA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005743 | mitochondrial inner membrane | - | TAS | Component |
| GO:0005783 | endoplasmic reticulum | 21873635. | IBA | Component |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005794 | Golgi apparatus | 21873635. | IBA | Component |
| GO:0005811 | lipid droplet | - | IDA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | 8381049. | NAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006644 | phospholipid metabolic process | - | TAS | Process |
| GO:0006654 | phosphatidic acid biosynthetic process | - | TAS | Process |
| GO:0006663 | platelet activating factor biosynthetic process | 9430701. | NAS | Process |
| GO:0006690 | icosanoid metabolic process | 9430701. | NAS | Process |
| GO:0019369 | arachidonic acid metabolic process | - | TAS | Process |
| GO:0031410 | cytoplasmic vesicle | - | IEA | Component |
| GO:0035965 | cardiolipin acyl-chain remodeling | - | TAS | Process |
| GO:0036148 | phosphatidylglycerol acyl-chain remodeling | - | TAS | Process |
| GO:0036149 | phosphatidylinositol acyl-chain remodeling | - | TAS | Process |
| GO:0036150 | phosphatidylserine acyl-chain remodeling | - | TAS | Process |
| GO:0036151 | phosphatidylcholine acyl-chain remodeling | - | TAS | Process |
| GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | - | TAS | Process |
| GO:0042127 | regulation of cell proliferation | - | IEA | Process |
| GO:0046456 | icosanoid biosynthetic process | - | IEA | Process |
| GO:0046475 | glycerophospholipid catabolic process | 21873635. | IBA | Process |
| GO:0047498 | calcium-dependent phospholipase A2 activity | 21873635. | IBA | Function |
| GO:0050482 | arachidonic acid secretion | - | IEA | Process |
| GO:0071236 | cellular response to antibiotic | - | IEA | Process |
| GO:0102567 | phospholipase A2 activity (consuming 1,2-dipalmitoylphosphatidylcholine) | - | IEA | Function |
| GO:0102568 | phospholipase A2 activity consuming 1,2-dioleoylphosphatidylethanolamine) | - | IEA | Function |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| MESO | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| KIRC | |||
| PAAD | |||
| READ | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| HNSC | ||
| BRCA | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| UVM | ||
| OV |