
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000310966 | RNA_polI_A34 | PF08208.11 | 1.6e-37 | 1 | 1 |
| ENSP00000465099 | RNA_polI_A34 | PF08208.11 | 1.6e-37 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 16809778 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000590794 | CD3EAP-203 | 489 | - | ENSP00000466503 | 67 (aa) | - | K7EMH2 |
| ENST00000309424 | CD3EAP-201 | 3286 | - | ENSP00000310966 | 510 (aa) | - | O15446 |
| ENST00000592852 | CD3EAP-204 | 1061 | - | ENSP00000467771 | 39 (aa) | - | K7EQC8 |
| ENST00000589804 | CD3EAP-202 | 1830 | - | ENSP00000465099 | 512 (aa) | - | O15446 |
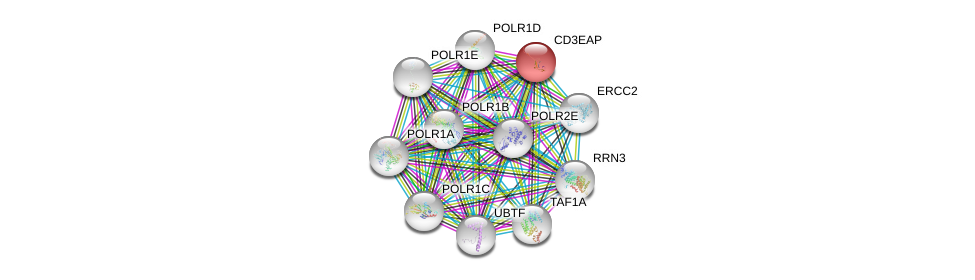
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000117877 | CD3EAP | 95 | 67.568 | ENSAMEG00000012438 | CD3EAP | 98 | 61.996 | Ailuropoda_melanoleuca |
| ENSG00000117877 | CD3EAP | 100 | 97.436 | ENSANAG00000022477 | CD3EAP | 100 | 86.019 | Aotus_nancymaae |
| ENSG00000117877 | CD3EAP | 99 | 65.537 | ENSBTAG00000023601 | CD3EAP | 98 | 65.779 | Bos_taurus |
| ENSG00000117877 | CD3EAP | 100 | 97.436 | ENSCJAG00000012689 | CD3EAP | 100 | 83.752 | Callithrix_jacchus |
| ENSG00000117877 | CD3EAP | 99 | 57.519 | ENSCAFG00000004455 | CD3EAP | 93 | 57.707 | Canis_familiaris |
| ENSG00000117877 | CD3EAP | 99 | 59.398 | ENSCAFG00020011342 | CD3EAP | 93 | 59.586 | Canis_lupus_dingo |
| ENSG00000117877 | CD3EAP | 99 | 65.537 | ENSCHIG00000022038 | CD3EAP | 97 | 65.970 | Capra_hircus |
| ENSG00000117877 | CD3EAP | 99 | 67.647 | ENSTSYG00000036564 | CD3EAP | 98 | 66.994 | Carlito_syrichta |
| ENSG00000117877 | CD3EAP | 97 | 60.853 | ENSCPOG00000033293 | CD3EAP | 98 | 60.687 | Cavia_porcellus |
| ENSG00000117877 | CD3EAP | 100 | 83.685 | ENSCCAG00000024253 | CD3EAP | 100 | 83.685 | Cebus_capucinus |
| ENSG00000117877 | CD3EAP | 99 | 91.016 | ENSCATG00000030677 | CD3EAP | 99 | 91.142 | Cercocebus_atys |
| ENSG00000117877 | CD3EAP | 97 | 58.189 | ENSCLAG00000004691 | CD3EAP | 87 | 57.736 | Chinchilla_lanigera |
| ENSG00000117877 | CD3EAP | 99 | 90.820 | ENSCSAG00000003181 | CD3EAP | 99 | 90.945 | Chlorocebus_sabaeus |
| ENSG00000117877 | CD3EAP | 99 | 90.039 | ENSCANG00000024271 | CD3EAP | 99 | 90.157 | Colobus_angolensis_palliatus |
| ENSG00000117877 | CD3EAP | 85 | 55.430 | ENSCGRG00001023584 | Cd3eap | 97 | 52.743 | Cricetulus_griseus_chok1gshd |
| ENSG00000117877 | CD3EAP | 99 | 62.949 | ENSDNOG00000041816 | CD3EAP | 98 | 62.998 | Dasypus_novemcinctus |
| ENSG00000117877 | CD3EAP | 57 | 59.871 | ENSDORG00000027052 | - | 87 | 56.704 | Dipodomys_ordii |
| ENSG00000117877 | CD3EAP | 98 | 66.348 | ENSEASG00005014188 | CD3EAP | 97 | 67.433 | Equus_asinus_asinus |
| ENSG00000117877 | CD3EAP | 98 | 66.348 | ENSECAG00000032490 | CD3EAP | 97 | 67.433 | Equus_caballus |
| ENSG00000117877 | CD3EAP | 70 | 58.824 | ENSEEUG00000012763 | - | 86 | 61.661 | Erinaceus_europaeus |
| ENSG00000117877 | CD3EAP | 99 | 58.974 | ENSFCAG00000043166 | CD3EAP | 99 | 58.974 | Felis_catus |
| ENSG00000117877 | CD3EAP | 82 | 57.412 | ENSFDAG00000012196 | - | 86 | 60.172 | Fukomys_damarensis |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSGGOG00000015872 | - | 100 | 97.070 | Gorilla_gorilla |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSGGOG00000041192 | - | 100 | 97.070 | Gorilla_gorilla |
| ENSG00000117877 | CD3EAP | 100 | 60.952 | ENSHGLG00000002795 | CD3EAP | 88 | 60.952 | Heterocephalus_glaber_female |
| ENSG00000117877 | CD3EAP | 100 | 60.952 | ENSHGLG00100006839 | CD3EAP | 88 | 60.952 | Heterocephalus_glaber_male |
| ENSG00000117877 | CD3EAP | 97 | 58.600 | ENSSTOG00000002799 | CD3EAP | 96 | 57.510 | Ictidomys_tridecemlineatus |
| ENSG00000117877 | CD3EAP | 83 | 53.647 | ENSJJAG00000013173 | Cd3eap | 87 | 55.833 | Jaculus_jaculus |
| ENSG00000117877 | CD3EAP | 95 | 59.459 | ENSLAFG00000001827 | CD3EAP | 84 | 69.359 | Loxodonta_africana |
| ENSG00000117877 | CD3EAP | 99 | 90.039 | ENSMFAG00000006908 | CD3EAP | 99 | 90.157 | Macaca_fascicularis |
| ENSG00000117877 | CD3EAP | 99 | 90.039 | ENSMNEG00000028086 | CD3EAP | 99 | 90.157 | Macaca_nemestrina |
| ENSG00000117877 | CD3EAP | 99 | 90.856 | ENSMLEG00000043530 | CD3EAP | 99 | 90.980 | Mandrillus_leucophaeus |
| ENSG00000117877 | CD3EAP | 66 | 60.116 | ENSMAUG00000007876 | Cd3eap | 86 | 58.960 | Mesocricetus_auratus |
| ENSG00000117877 | CD3EAP | 99 | 67.954 | ENSMICG00000031258 | CD3EAP | 98 | 67.057 | Microcebus_murinus |
| ENSG00000117877 | CD3EAP | 66 | 59.347 | ENSMOCG00000013766 | Cd3eap | 82 | 59.880 | Microtus_ochrogaster |
| ENSG00000117877 | CD3EAP | 81 | 33.864 | ENSMODG00000021547 | - | 81 | 39.286 | Monodelphis_domestica |
| ENSG00000117877 | CD3EAP | 82 | 53.882 | MGP_CAROLIEiJ_G0029285 | Cd3eap | 81 | 57.692 | Mus_caroli |
| ENSG00000117877 | CD3EAP | 84 | 54.756 | ENSMUSG00000047649 | Cd3eap | 83 | 58.382 | Mus_musculus |
| ENSG00000117877 | CD3EAP | 84 | 54.608 | MGP_SPRETEiJ_G0030369 | Cd3eap | 97 | 54.654 | Mus_spretus |
| ENSG00000117877 | CD3EAP | 99 | 61.260 | ENSMPUG00000005965 | CD3EAP | 99 | 61.641 | Mustela_putorius_furo |
| ENSG00000117877 | CD3EAP | 95 | 75.676 | ENSMLUG00000012319 | CD3EAP | 97 | 64.567 | Myotis_lucifugus |
| ENSG00000117877 | CD3EAP | 77 | 66.667 | ENSNGAG00000015313 | Cd3eap | 88 | 61.236 | Nannospalax_galili |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSNLEG00000035363 | CD3EAP | 100 | 94.902 | Nomascus_leucogenys |
| ENSG00000117877 | CD3EAP | 83 | 59.719 | ENSODEG00000007587 | - | 99 | 60.190 | Octodon_degus |
| ENSG00000117877 | CD3EAP | 97 | 67.659 | ENSOGAG00000013782 | CD3EAP | 99 | 66.602 | Otolemur_garnettii |
| ENSG00000117877 | CD3EAP | 99 | 65.160 | ENSOARG00000009943 | CD3EAP | 98 | 65.589 | Ovis_aries |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSPPAG00000004958 | CD3EAP | 100 | 98.828 | Pan_paniscus |
| ENSG00000117877 | CD3EAP | 99 | 59.055 | ENSPPRG00000001954 | CD3EAP | 99 | 59.449 | Panthera_pardus |
| ENSG00000117877 | CD3EAP | 74 | 68.966 | ENSPTIG00000014078 | - | 97 | 58.652 | Panthera_tigris_altaica |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSPTRG00000011151 | CD3EAP | 100 | 99.022 | Pan_troglodytes |
| ENSG00000117877 | CD3EAP | 99 | 91.634 | ENSPANG00000014599 | CD3EAP | 99 | 91.765 | Papio_anubis |
| ENSG00000117877 | CD3EAP | 97 | 63.158 | ENSPEMG00000018578 | Cd3eap | 82 | 63.050 | Peromyscus_maniculatus_bairdii |
| ENSG00000117877 | CD3EAP | 54 | 40.214 | ENSPCIG00000022939 | - | 63 | 48.000 | Phascolarctos_cinereus |
| ENSG00000117877 | CD3EAP | 100 | 100.000 | ENSPPYG00000010119 | CD3EAP | 99 | 95.635 | Pongo_abelii |
| ENSG00000117877 | CD3EAP | 97 | 65.789 | ENSPCAG00000012953 | - | 99 | 66.129 | Procavia_capensis |
| ENSG00000117877 | CD3EAP | 100 | 68.511 | ENSPCOG00000028780 | CD3EAP | 98 | 68.605 | Propithecus_coquereli |
| ENSG00000117877 | CD3EAP | 99 | 66.139 | ENSPVAG00000001330 | CD3EAP | 97 | 67.002 | Pteropus_vampyrus |
| ENSG00000117877 | CD3EAP | 94 | 53.608 | ENSRNOG00000016825 | Cd3eap | 100 | 52.429 | Rattus_norvegicus |
| ENSG00000117877 | CD3EAP | 99 | 90.625 | ENSRBIG00000040738 | CD3EAP | 99 | 90.748 | Rhinopithecus_bieti |
| ENSG00000117877 | CD3EAP | 99 | 90.802 | ENSRROG00000043410 | CD3EAP | 99 | 90.927 | Rhinopithecus_roxellana |
| ENSG00000117877 | CD3EAP | 100 | 80.851 | ENSSBOG00000032782 | CD3EAP | 100 | 81.044 | Saimiri_boliviensis_boliviensis |
| ENSG00000117877 | CD3EAP | 68 | 53.258 | ENSSARG00000008700 | - | 90 | 56.678 | Sorex_araneus |
| ENSG00000117877 | CD3EAP | 95 | 70.270 | ENSSSCG00000020754 | CD3EAP | 98 | 63.105 | Sus_scrofa |
| ENSG00000117877 | CD3EAP | 99 | 64.368 | ENSTTRG00000008311 | CD3EAP | 97 | 65.815 | Tursiops_truncatus |
| ENSG00000117877 | CD3EAP | 99 | 62.311 | ENSUAMG00000020204 | CD3EAP | 98 | 62.380 | Ursus_americanus |
| ENSG00000117877 | CD3EAP | 99 | 62.311 | ENSUMAG00000023587 | CD3EAP | 98 | 62.188 | Ursus_maritimus |
| ENSG00000117877 | CD3EAP | 99 | 57.491 | ENSVVUG00000004115 | CD3EAP | 93 | 58.083 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000120 | RNA polymerase I transcription factor complex | 9426281. | TAS | Component |
| GO:0001650 | fibrillar center | - | IDA | Component |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0003899 | DNA-directed 5'-3' RNA polymerase activity | - | IEA | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005694 | chromosome | - | IEA | Component |
| GO:0005730 | nucleolus | 9426281. | TAS | Component |
| GO:0005739 | mitochondrion | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006361 | transcription initiation from RNA polymerase I promoter | - | TAS | Process |
| GO:0006362 | transcription elongation from RNA polymerase I promoter | - | TAS | Process |
| GO:0006363 | termination of RNA polymerase I transcription | - | TAS | Process |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 10373416. | TAS | Process |
| GO:0009303 | rRNA transcription | 9426281. | TAS | Process |
| GO:0045815 | positive regulation of gene expression, epigenetic | - | TAS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRP | |||
| READ | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| HNSC | ||
| SKCM | ||
| KIRP | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| THCA | ||
| LUAD | ||
| UVM |