
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000400107 | MMR_HSR1 | PF01926.23 | 0.0002 | 1 | 1 |
| ENSP00000362946 | MMR_HSR1 | PF01926.23 | 0.0003 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000373840 | RAB14-201 | 4145 | - | ENSP00000362946 | 215 (aa) | - | P61106 |
| ENST00000451303 | RAB14-202 | 711 | - | ENSP00000400107 | 181 (aa) | - | X6RFL8 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04152 | AMPK signaling pathway | KEGG |
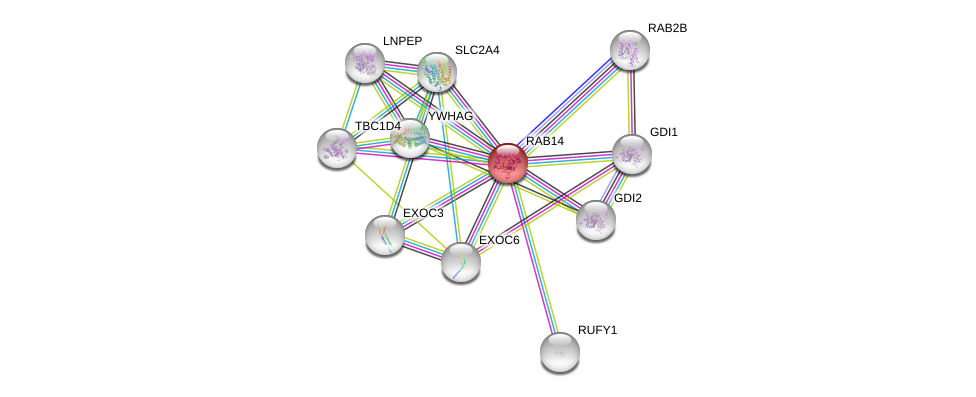
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000119396 | RAB14 | 88 | 39.394 | ENSG00000075785 | RAB7A | 91 | 39.416 |
| ENSG00000119396 | RAB14 | 89 | 32.727 | ENSG00000132341 | RAN | 83 | 30.994 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSAPOG00000008641 | rab14 | 100 | 100.000 | Acanthochromis_polyacanthus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSAOCG00000008492 | rab14 | 100 | 100.000 | Amphiprion_ocellaris |
| ENSG00000119396 | RAB14 | 100 | 95.580 | ENSAOCG00000018191 | - | 100 | 93.023 | Amphiprion_ocellaris |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSAPEG00000014246 | rab14 | 100 | 100.000 | Amphiprion_percula |
| ENSG00000119396 | RAB14 | 100 | 95.580 | ENSAPEG00000022217 | - | 100 | 93.023 | Amphiprion_percula |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSATEG00000018600 | rab14 | 100 | 100.000 | Anabas_testudineus |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSATEG00000012604 | - | 100 | 93.488 | Anabas_testudineus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSACAG00000008285 | RAB14 | 100 | 100.000 | Anolis_carolinensis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSANAG00000031127 | RAB14 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSACLG00000006329 | - | 100 | 92.093 | Astatotilapia_calliptera |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSACLG00000013505 | - | 100 | 99.070 | Astatotilapia_calliptera |
| ENSG00000119396 | RAB14 | 100 | 99.535 | ENSAMXG00000040126 | - | 79 | 99.535 | Astyanax_mexicanus |
| ENSG00000119396 | RAB14 | 100 | 98.895 | ENSAMXG00000009640 | rab14 | 100 | 96.279 | Astyanax_mexicanus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSBTAG00000017803 | RAB14 | 100 | 100.000 | Bos_taurus |
| ENSG00000119396 | RAB14 | 100 | 91.713 | WBGene00004276 | rab-14 | 100 | 82.791 | Caenorhabditis_elegans |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCJAG00000042020 | RAB14 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCAFG00000003629 | RAB14 | 100 | 100.000 | Canis_familiaris |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCAFG00020009287 | RAB14 | 100 | 100.000 | Canis_lupus_dingo |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCHIG00000026983 | RAB14 | 100 | 100.000 | Capra_hircus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSTSYG00000014136 | - | 100 | 100.000 | Carlito_syrichta |
| ENSG00000119396 | RAB14 | 79 | 93.007 | ENSCAPG00000017256 | RAB14 | 100 | 93.007 | Cavia_aperea |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCPOG00000036631 | RAB14 | 100 | 100.000 | Cavia_porcellus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCCAG00000020267 | RAB14 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCATG00000035718 | RAB14 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCLAG00000009500 | RAB14 | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCSAG00000013289 | RAB14 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCPBG00000009941 | RAB14 | 100 | 100.000 | Chrysemys_picta_bellii |
| ENSG00000119396 | RAB14 | 100 | 94.475 | ENSCING00000006942 | - | 100 | 89.302 | Ciona_intestinalis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCANG00000039448 | RAB14 | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCGRG00001006440 | Rab14 | 100 | 100.000 | Cricetulus_griseus_chok1gshd |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCGRG00000005328 | Rab14 | 100 | 100.000 | Cricetulus_griseus_crigri |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCSEG00000016498 | rab14 | 100 | 99.535 | Cynoglossus_semilaevis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSCVAG00000022353 | rab14 | 100 | 100.000 | Cyprinodon_variegatus |
| ENSG00000119396 | RAB14 | 100 | 98.895 | ENSDARG00000074246 | rab14 | 100 | 96.279 | Danio_rerio |
| ENSG00000119396 | RAB14 | 100 | 99.448 | ENSDARG00000035188 | rab14l | 100 | 99.070 | Danio_rerio |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSDORG00000023851 | Rab14 | 100 | 100.000 | Dipodomys_ordii |
| ENSG00000119396 | RAB14 | 100 | 86.740 | FBgn0015791 | Rab14 | 100 | 81.395 | Drosophila_melanogaster |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSEASG00005021919 | RAB14 | 100 | 100.000 | Equus_asinus_asinus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSECAG00000011518 | RAB14 | 100 | 100.000 | Equus_caballus |
| ENSG00000119396 | RAB14 | 81 | 100.000 | ENSEEUG00000013074 | RAB14 | 100 | 79.535 | Erinaceus_europaeus |
| ENSG00000119396 | RAB14 | 100 | 98.895 | ENSELUG00000009832 | rab14 | 100 | 98.140 | Esox_lucius |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSELUG00000013571 | - | 100 | 100.000 | Esox_lucius |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSFCAG00000026542 | RAB14 | 100 | 100.000 | Felis_catus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSFDAG00000007443 | RAB14 | 100 | 100.000 | Fukomys_damarensis |
| ENSG00000119396 | RAB14 | 81 | 100.000 | ENSFHEG00000018450 | RAB14 | 75 | 100.000 | Fundulus_heteroclitus |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSFHEG00000000695 | - | 100 | 93.953 | Fundulus_heteroclitus |
| ENSG00000119396 | RAB14 | 100 | 94.475 | ENSGMOG00000000532 | - | 100 | 92.093 | Gadus_morhua |
| ENSG00000119396 | RAB14 | 86 | 98.718 | ENSGMOG00000011766 | - | 100 | 98.718 | Gadus_morhua |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSGALG00000001474 | RAB14 | 100 | 100.000 | Gallus_gallus |
| ENSG00000119396 | RAB14 | 95 | 100.000 | ENSGACG00000016799 | - | 100 | 100.000 | Gasterosteus_aculeatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSGAGG00000004988 | RAB14 | 100 | 100.000 | Gopherus_agassizii |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSGGOG00000027624 | RAB14 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSHBUG00000022562 | - | 100 | 99.070 | Haplochromis_burtoni |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSHBUG00000006056 | - | 100 | 92.093 | Haplochromis_burtoni |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSHGLG00000013486 | RAB14 | 100 | 100.000 | Heterocephalus_glaber_female |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSHGLG00100010554 | RAB14 | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000119396 | RAB14 | 100 | 95.028 | ENSHCOG00000013180 | - | 100 | 92.093 | Hippocampus_comes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSHCOG00000019224 | rab14 | 100 | 100.000 | Hippocampus_comes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSIPUG00000015525 | - | 100 | 98.605 | Ictalurus_punctatus |
| ENSG00000119396 | RAB14 | 100 | 98.895 | ENSIPUG00000007415 | rab14 | 100 | 96.279 | Ictalurus_punctatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSTOG00000025248 | RAB14 | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSJJAG00000012923 | - | 100 | 100.000 | Jaculus_jaculus |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSKMAG00000011301 | - | 100 | 93.953 | Kryptolebias_marmoratus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSKMAG00000017487 | rab14 | 100 | 100.000 | Kryptolebias_marmoratus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSLBEG00000019435 | rab14 | 100 | 100.000 | Labrus_bergylta |
| ENSG00000119396 | RAB14 | 100 | 95.580 | ENSLBEG00000026790 | - | 100 | 93.023 | Labrus_bergylta |
| ENSG00000119396 | RAB14 | 100 | 61.395 | ENSLBEG00000015149 | - | 100 | 61.395 | Labrus_bergylta |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSLACG00000011181 | RAB14 | 100 | 100.000 | Latimeria_chalumnae |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSLOCG00000005509 | rab14 | 100 | 100.000 | Lepisosteus_oculatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSLAFG00000005250 | RAB14 | 100 | 100.000 | Loxodonta_africana |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMFAG00000046157 | RAB14 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMMUG00000047957 | RAB14 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMNEG00000036774 | RAB14 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMLEG00000031858 | RAB14 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSMAMG00000018450 | - | 100 | 93.488 | Mastacembelus_armatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMAMG00000012750 | rab14 | 100 | 100.000 | Mastacembelus_armatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMZEG00005018759 | - | 100 | 99.070 | Maylandia_zebra |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSMZEG00005001885 | - | 100 | 92.093 | Maylandia_zebra |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMGAG00000007763 | RAB14 | 100 | 100.000 | Meleagris_gallopavo |
| ENSG00000119396 | RAB14 | 81 | 100.000 | ENSMAUG00000014779 | Rab14 | 98 | 100.000 | Mesocricetus_auratus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMICG00000003350 | RAB14 | 100 | 100.000 | Microcebus_murinus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMOCG00000003760 | Rab14 | 100 | 100.000 | Microtus_ochrogaster |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMODG00000019649 | RAB14 | 100 | 100.000 | Monodelphis_domestica |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSMALG00000003956 | - | 100 | 93.488 | Monopterus_albus |
| ENSG00000119396 | RAB14 | 100 | 86.512 | ENSMALG00000022118 | rab14 | 100 | 86.512 | Monopterus_albus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | MGP_CAROLIEiJ_G0023477 | - | 100 | 100.000 | Mus_caroli |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMUSG00000026878 | Rab14 | 100 | 100.000 | Mus_musculus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | MGP_PahariEiJ_G0024934 | Rab14 | 100 | 100.000 | Mus_pahari |
| ENSG00000119396 | RAB14 | 100 | 100.000 | MGP_SPRETEiJ_G0024394 | Rab14 | 100 | 100.000 | Mus_spretus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMPUG00000007533 | RAB14 | 100 | 100.000 | Mustela_putorius_furo |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSMLUG00000009818 | RAB14 | 100 | 100.000 | Myotis_lucifugus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSNGAG00000010962 | Rab14 | 100 | 100.000 | Nannospalax_galili |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSNBRG00000005542 | - | 100 | 99.070 | Neolamprologus_brichardi |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSNBRG00000002726 | - | 100 | 92.093 | Neolamprologus_brichardi |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSNLEG00000017108 | RAB14 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000119396 | RAB14 | 100 | 99.070 | ENSOPRG00000016426 | RAB14 | 100 | 99.070 | Ochotona_princeps |
| ENSG00000119396 | RAB14 | 100 | 99.535 | ENSODEG00000002013 | RAB14 | 100 | 99.535 | Octodon_degus |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSONIG00000000558 | - | 100 | 92.093 | Oreochromis_niloticus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSONIG00000014099 | rab14 | 100 | 99.070 | Oreochromis_niloticus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSOANG00000001101 | RAB14 | 100 | 100.000 | Ornithorhynchus_anatinus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSOCUG00000013654 | RAB14 | 100 | 100.000 | Oryctolagus_cuniculus |
| ENSG00000119396 | RAB14 | 100 | 95.028 | ENSORLG00000029921 | - | 100 | 92.093 | Oryzias_latipes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSORLG00000007406 | rab14 | 100 | 100.000 | Oryzias_latipes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSORLG00020018986 | rab14 | 100 | 95.349 | Oryzias_latipes_hni |
| ENSG00000119396 | RAB14 | 100 | 95.028 | ENSORLG00015000893 | - | 100 | 92.093 | Oryzias_latipes_hsok |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSORLG00015006881 | rab14 | 100 | 100.000 | Oryzias_latipes_hsok |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSOMEG00000003623 | rab14 | 100 | 100.000 | Oryzias_melastigma |
| ENSG00000119396 | RAB14 | 100 | 95.028 | ENSOMEG00000009290 | - | 100 | 91.628 | Oryzias_melastigma |
| ENSG00000119396 | RAB14 | 100 | 96.279 | ENSOGAG00000012973 | RAB14 | 100 | 96.279 | Otolemur_garnettii |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSOARG00000005498 | RAB14 | 100 | 100.000 | Ovis_aries |
| ENSG00000119396 | RAB14 | 100 | 93.548 | ENSPPAG00000031759 | RAB14 | 100 | 93.548 | Pan_paniscus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPPRG00000021393 | RAB14 | 100 | 100.000 | Panthera_pardus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPTIG00000017871 | RAB14 | 100 | 100.000 | Panthera_tigris_altaica |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPTRG00000021312 | RAB14 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPANG00000025819 | RAB14 | 100 | 100.000 | Papio_anubis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPKIG00000006906 | rab14 | 100 | 99.535 | Paramormyrops_kingsleyae |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPKIG00000004170 | - | 100 | 99.070 | Paramormyrops_kingsleyae |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPSIG00000003919 | RAB14 | 100 | 100.000 | Pelodiscus_sinensis |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSPMAG00000002006 | - | 100 | 92.093 | Petromyzon_marinus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPCIG00000009235 | RAB14 | 100 | 100.000 | Phascolarctos_cinereus |
| ENSG00000119396 | RAB14 | 100 | 99.537 | ENSPFOG00000019821 | RAB14 | 100 | 99.537 | Poecilia_formosa |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSPFOG00000001167 | - | 100 | 93.953 | Poecilia_formosa |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPLAG00000004551 | - | 100 | 100.000 | Poecilia_latipinna |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSPLAG00000022515 | - | 100 | 93.953 | Poecilia_latipinna |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPMEG00000010857 | rab14 | 100 | 100.000 | Poecilia_mexicana |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSPMEG00000006424 | - | 100 | 93.953 | Poecilia_mexicana |
| ENSG00000119396 | RAB14 | 100 | 96.154 | ENSPREG00000017344 | - | 100 | 93.519 | Poecilia_reticulata |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPCOG00000017682 | RAB14 | 100 | 100.000 | Propithecus_coquereli |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSPNYG00000005326 | - | 100 | 92.093 | Pundamilia_nyererei |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPNYG00000007314 | - | 100 | 99.070 | Pundamilia_nyererei |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSPNAG00000014363 | - | 100 | 100.000 | Pygocentrus_nattereri |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSRNOG00000018901 | Rab14 | 100 | 100.000 | Rattus_norvegicus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSRBIG00000036040 | RAB14 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSRROG00000038344 | RAB14 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSBOG00000027362 | RAB14 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSHAG00000017800 | RAB14 | 100 | 100.000 | Sarcophilus_harrisii |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSFOG00015011318 | - | 100 | 99.535 | Scleropages_formosus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSFOG00015012785 | rab14 | 100 | 99.535 | Scleropages_formosus |
| ENSG00000119396 | RAB14 | 100 | 95.580 | ENSSMAG00000018454 | - | 100 | 93.023 | Scophthalmus_maximus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSMAG00000003166 | rab14 | 100 | 100.000 | Scophthalmus_maximus |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSSDUG00000013050 | - | 100 | 93.488 | Seriola_dumerili |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSDUG00000004551 | rab14 | 100 | 100.000 | Seriola_dumerili |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSLDG00000010503 | rab14 | 100 | 100.000 | Seriola_lalandi_dorsalis |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSSLDG00000005718 | - | 100 | 93.488 | Seriola_lalandi_dorsalis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSPUG00000018803 | RAB14 | 94 | 99.535 | Sphenodon_punctatus |
| ENSG00000119396 | RAB14 | 100 | 95.580 | ENSSPAG00000005141 | - | 100 | 93.023 | Stegastes_partitus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSPAG00000022927 | rab14 | 100 | 100.000 | Stegastes_partitus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSSSCG00000005514 | RAB14 | 100 | 100.000 | Sus_scrofa |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSTGUG00000006657 | RAB14 | 100 | 100.000 | Taeniopygia_guttata |
| ENSG00000119396 | RAB14 | 100 | 96.133 | ENSTRUG00000011399 | - | 100 | 93.023 | Takifugu_rubripes |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSTNIG00000009039 | rab14 | 98 | 100.000 | Tetraodon_nigroviridis |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSTTRG00000010425 | RAB14 | 100 | 100.000 | Tursiops_truncatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSUAMG00000003164 | RAB14 | 100 | 100.000 | Ursus_americanus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSUMAG00000003596 | RAB14 | 100 | 100.000 | Ursus_maritimus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSVPAG00000009203 | RAB14 | 100 | 100.000 | Vicugna_pacos |
| ENSG00000119396 | RAB14 | 100 | 96.685 | ENSXMAG00000015075 | - | 100 | 93.953 | Xiphophorus_maculatus |
| ENSG00000119396 | RAB14 | 100 | 100.000 | ENSXMAG00000005294 | - | 100 | 100.000 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | - | IEA | Component |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0003924 | GTPase activity | 16034420.17562788. | IDA | Function |
| GO:0003924 | GTPase activity | 15004230. | NAS | Function |
| GO:0005515 | protein binding | 16034420.16237761.16902405.25416956. | IPI | Function |
| GO:0005525 | GTP binding | 16034420. | IDA | Function |
| GO:0005764 | lysosome | - | ISS | Component |
| GO:0005765 | lysosomal membrane | 17897319. | HDA | Component |
| GO:0005769 | early endosome | 21873635. | IBA | Component |
| GO:0005769 | early endosome | - | ISS | Component |
| GO:0005770 | late endosome | - | ISS | Component |
| GO:0005791 | rough endoplasmic reticulum | - | ISS | Component |
| GO:0005795 | Golgi stack | - | ISS | Component |
| GO:0005802 | trans-Golgi network | - | IEA | Component |
| GO:0005829 | cytosol | - | ISS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | ISS | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005929 | cilium | 17646400. | IDA | Component |
| GO:0006661 | phosphatidylinositol biosynthetic process | - | TAS | Process |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0006895 | Golgi to endosome transport | 21873635. | IBA | Process |
| GO:0006895 | Golgi to endosome transport | - | ISS | Process |
| GO:0006895 | Golgi to endosome transport | 15004230. | TAS | Process |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | - | ISS | Process |
| GO:0016192 | vesicle-mediated transport | 15004230. | NAS | Process |
| GO:0019003 | GDP binding | 16034420. | IDA | Function |
| GO:0030140 | trans-Golgi network transport vesicle | - | ISS | Component |
| GO:0031489 | myosin V binding | 24006491. | IPI | Function |
| GO:0031901 | early endosome membrane | - | TAS | Component |
| GO:0032456 | endocytic recycling | 22595670. | IDA | Process |
| GO:0032482 | Rab protein signal transduction | 21873635. | IBA | Process |
| GO:0032880 | regulation of protein localization | 22595670. | IDA | Process |
| GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network | - | ISS | Component |
| GO:0042742 | defense response to bacterium | - | IEA | Process |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0045335 | phagocytic vesicle | 21873635. | IBA | Component |
| GO:0045335 | phagocytic vesicle | 21255211. | IDA | Component |
| GO:0045995 | regulation of embryonic development | - | ISS | Process |
| GO:0046907 | intracellular transport | 15004230. | NAS | Process |
| GO:0048471 | perinuclear region of cytoplasm | - | ISS | Component |
| GO:0055037 | recycling endosome | 22595670. | IDA | Component |
| GO:0055038 | recycling endosome membrane | - | TAS | Component |
| GO:0070062 | extracellular exosome | 19056867.20458337. | HDA | Component |
| GO:0070821 | tertiary granule membrane | - | TAS | Component |
| GO:0090387 | phagolysosome assembly involved in apoptotic cell clearance | 21873635. | IBA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CHOL | |||
| LAML | |||
| LIHC | |||
| PRAD | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| UCS | ||
| PRAD | ||
| BRCA | ||
| KIRP | ||
| PCPG | ||
| LAML | ||
| LGG |