
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000349250 | MMR_HSR1 | PF01926.23 | 4.7e-07 | 1 | 1 |
| ENSP00000379897 | MMR_HSR1 | PF01926.23 | 4.7e-07 | 1 | 1 |
| ENSP00000379898 | MMR_HSR1 | PF01926.23 | 4.7e-07 | 1 | 1 |
| ENSP00000379899 | MMR_HSR1 | PF01926.23 | 4.7e-07 | 1 | 1 |
| ENSP00000385236 | MMR_HSR1 | PF01926.23 | 4.7e-07 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000356797 | ARL4A-201 | 1147 | - | ENSP00000349250 | 200 (aa) | - | P40617 |
| ENST00000404894 | ARL4A-205 | 840 | - | ENSP00000385236 | 200 (aa) | - | P40617 |
| ENST00000396664 | ARL4A-204 | 1144 | - | ENSP00000379899 | 200 (aa) | - | P40617 |
| ENST00000439721 | ARL4A-206 | 845 | - | ENSP00000397651 | 50 (aa) | - | C9J7Q9 |
| ENST00000396663 | ARL4A-203 | 3212 | - | ENSP00000379898 | 200 (aa) | - | P40617 |
| ENST00000396662 | ARL4A-202 | 1296 | - | ENSP00000379897 | 200 (aa) | - | P40617 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000122644 | Iron | 1.0660000E-004 | 19880490 |
| ENSG00000122644 | Iron | 7.0390000E-004 | 19880490 |
| ENSG00000122644 | Iron | 9.0080000E-005 | 19880490 |
| ENSG00000122644 | Child Development Disorders, Pervasive | 1.8629000E-005 | - |
| ENSG00000122644 | Alcoholism | 8.4900000E-008 | - |
| ENSG00000122644 | Mental Disorders | 5E-6 | 20889312 |
| ENSG00000122644 | Death, Sudden, Cardiac | 4E-6 | 21658281 |
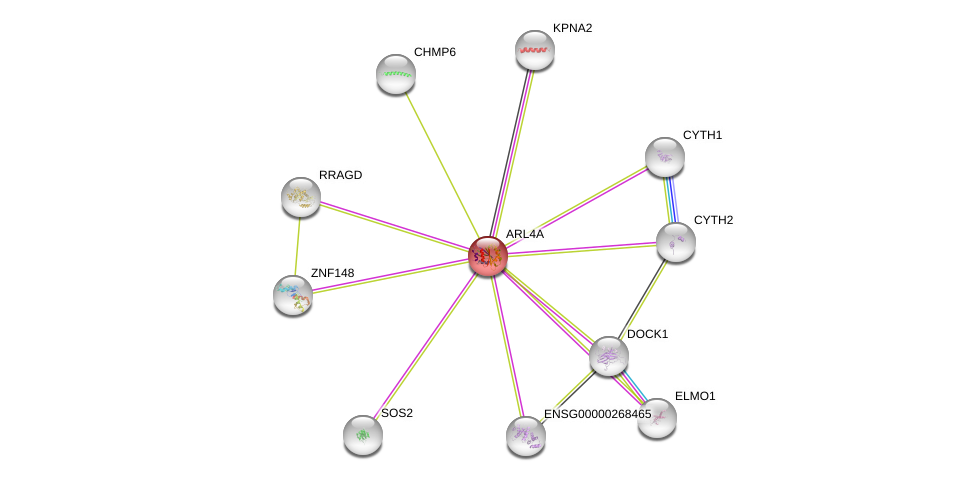
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000122644 | ARL4A | 95 | 81.675 | ENSAPOG00000010415 | - | 95 | 81.675 | Acanthochromis_polyacanthus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSAMEG00000019250 | ARL4A | 100 | 99.500 | Ailuropoda_melanoleuca |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSACIG00000009110 | arl4aa | 100 | 81.500 | Amphilophus_citrinellus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSACIG00000000949 | - | 100 | 81.095 | Amphilophus_citrinellus |
| ENSG00000122644 | ARL4A | 92 | 83.784 | ENSAOCG00000018628 | - | 100 | 83.784 | Amphiprion_ocellaris |
| ENSG00000122644 | ARL4A | 100 | 81.000 | ENSAPEG00000000558 | arl4aa | 100 | 81.000 | Amphiprion_percula |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSAPEG00000017251 | - | 100 | 83.085 | Amphiprion_percula |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSATEG00000003663 | arl4aa | 99 | 75.377 | Anabas_testudineus |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSAPLG00000001404 | ARL4A | 100 | 96.000 | Anas_platyrhynchos |
| ENSG00000122644 | ARL4A | 100 | 95.000 | ENSACAG00000013615 | ARL4A | 100 | 95.000 | Anolis_carolinensis |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSANAG00000018721 | - | 100 | 98.000 | Aotus_nancymaae |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSACLG00000016659 | - | 100 | 80.597 | Astatotilapia_calliptera |
| ENSG00000122644 | ARL4A | 91 | 80.874 | ENSAMXG00000025423 | arl4ab | 96 | 80.874 | Astyanax_mexicanus |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSAMXG00000006638 | arl4aa | 96 | 84.896 | Astyanax_mexicanus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSBTAG00000002869 | ARL4A | 100 | 99.500 | Bos_taurus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSCAFG00000002318 | ARL4A | 100 | 99.500 | Canis_familiaris |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSCHIG00000011984 | ARL4A | 100 | 99.500 | Capra_hircus |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSCPOG00000000543 | ARL4A | 100 | 98.000 | Cavia_porcellus |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSCATG00000016895 | - | 93 | 98.500 | Cercocebus_atys |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSCATG00000022471 | - | 100 | 100.000 | Cercocebus_atys |
| ENSG00000122644 | ARL4A | 93 | 79.032 | ENSCLAG00000008391 | - | 94 | 76.000 | Chinchilla_lanigera |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSCLAG00000006259 | ARL4A | 100 | 98.000 | Chinchilla_lanigera |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSCSAG00000018752 | - | 100 | 96.000 | Chlorocebus_sabaeus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSCSAG00000019116 | - | 100 | 99.500 | Chlorocebus_sabaeus |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSCPBG00000024120 | ARL4A | 100 | 96.000 | Chrysemys_picta_bellii |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSCANG00000001658 | ARL4A | 100 | 99.500 | Colobus_angolensis_palliatus |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSCGRG00001004637 | Arl4a | 100 | 98.500 | Cricetulus_griseus_chok1gshd |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSCGRG00000009701 | Arl4a | 100 | 98.500 | Cricetulus_griseus_crigri |
| ENSG00000122644 | ARL4A | 100 | 86.000 | ENSCSEG00000007479 | ARL4A | 100 | 81.500 | Cynoglossus_semilaevis |
| ENSG00000122644 | ARL4A | 98 | 80.102 | ENSCSEG00000014890 | arl4aa | 100 | 80.102 | Cynoglossus_semilaevis |
| ENSG00000122644 | ARL4A | 100 | 78.000 | ENSCSEG00000010194 | - | 98 | 71.574 | Cynoglossus_semilaevis |
| ENSG00000122644 | ARL4A | 100 | 81.500 | ENSCVAG00000004305 | arl4aa | 100 | 81.500 | Cyprinodon_variegatus |
| ENSG00000122644 | ARL4A | 100 | 80.100 | ENSDARG00000033182 | arl4ab | 100 | 80.100 | Danio_rerio |
| ENSG00000122644 | ARL4A | 100 | 78.000 | ENSDARG00000099918 | arl4aa | 100 | 79.000 | Danio_rerio |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSDNOG00000011303 | ARL4A | 100 | 99.500 | Dasypus_novemcinctus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSDORG00000028437 | Arl4a | 100 | 99.500 | Dipodomys_ordii |
| ENSG00000122644 | ARL4A | 91 | 59.116 | ENSEBUG00000005056 | arl4aa | 84 | 59.116 | Eptatretus_burgeri |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSECAG00000019333 | ARL4A | 100 | 98.000 | Equus_caballus |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSELUG00000023460 | - | 99 | 75.490 | Esox_lucius |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSFCAG00000035870 | ARL4A | 100 | 99.500 | Felis_catus |
| ENSG00000122644 | ARL4A | 100 | 97.500 | ENSFALG00000014498 | ARL4A | 100 | 97.500 | Ficedula_albicollis |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSFDAG00000019045 | ARL4A | 100 | 98.000 | Fukomys_damarensis |
| ENSG00000122644 | ARL4A | 100 | 81.000 | ENSFHEG00000009487 | arl4aa | 100 | 81.000 | Fundulus_heteroclitus |
| ENSG00000122644 | ARL4A | 100 | 72.000 | ENSGMOG00000020530 | arl4aa | 100 | 71.569 | Gadus_morhua |
| ENSG00000122644 | ARL4A | 100 | 96.500 | ENSGALG00000027757 | ARL4A | 100 | 96.500 | Gallus_gallus |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSGAFG00000014283 | arl4aa | 100 | 79.500 | Gambusia_affinis |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSGACG00000000497 | - | 100 | 78.607 | Gasterosteus_aculeatus |
| ENSG00000122644 | ARL4A | 100 | 81.000 | ENSGACG00000012810 | arl4aa | 100 | 81.000 | Gasterosteus_aculeatus |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSGAGG00000019544 | ARL4A | 100 | 96.000 | Gopherus_agassizii |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSGGOG00000044019 | ARL4A | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSGGOG00000039058 | - | 91 | 95.980 | Gorilla_gorilla |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSGGOG00000006954 | - | 90 | 90.526 | Gorilla_gorilla |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSHBUG00000012803 | arl4aa | 100 | 81.500 | Haplochromis_burtoni |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSHBUG00000007873 | - | 100 | 80.597 | Haplochromis_burtoni |
| ENSG00000122644 | ARL4A | 100 | 97.500 | ENSHGLG00000006478 | ARL4A | 100 | 97.500 | Heterocephalus_glaber_female |
| ENSG00000122644 | ARL4A | 100 | 97.500 | ENSHGLG00100008548 | ARL4A | 100 | 97.500 | Heterocephalus_glaber_male |
| ENSG00000122644 | ARL4A | 100 | 78.000 | ENSHCOG00000014829 | - | 99 | 80.100 | Hippocampus_comes |
| ENSG00000122644 | ARL4A | 96 | 78.010 | ENSHCOG00000006452 | arl4aa | 99 | 78.010 | Hippocampus_comes |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSIPUG00000024159 | arl4a | 100 | 82.412 | Ictalurus_punctatus |
| ENSG00000122644 | ARL4A | 96 | 79.688 | ENSIPUG00000021618 | arl4ab | 99 | 79.688 | Ictalurus_punctatus |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSSTOG00000005161 | ARL4A | 100 | 99.000 | Ictidomys_tridecemlineatus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSJJAG00000011436 | - | 100 | 99.500 | Jaculus_jaculus |
| ENSG00000122644 | ARL4A | 100 | 92.000 | ENSJJAG00000011618 | - | 100 | 92.000 | Jaculus_jaculus |
| ENSG00000122644 | ARL4A | 94 | 81.915 | ENSKMAG00000020210 | arl4aa | 97 | 81.915 | Kryptolebias_marmoratus |
| ENSG00000122644 | ARL4A | 100 | 86.000 | ENSLBEG00000013638 | arl4aa | 98 | 82.222 | Labrus_bergylta |
| ENSG00000122644 | ARL4A | 100 | 92.000 | ENSLACG00000002695 | ARL4A | 97 | 90.500 | Latimeria_chalumnae |
| ENSG00000122644 | ARL4A | 100 | 88.000 | ENSLOCG00000018349 | arl4aa | 100 | 87.000 | Lepisosteus_oculatus |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSLAFG00000022094 | ARL4A | 100 | 99.000 | Loxodonta_africana |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSMFAG00000045072 | - | 100 | 84.000 | Macaca_fascicularis |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMFAG00000008326 | - | 100 | 99.500 | Macaca_fascicularis |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMMUG00000010846 | ARL4A | 100 | 99.500 | Macaca_mulatta |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSMMUG00000042759 | - | 100 | 98.000 | Macaca_mulatta |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSMMUG00000014576 | - | 100 | 98.500 | Macaca_mulatta |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSMNEG00000026898 | - | 100 | 98.500 | Macaca_nemestrina |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMNEG00000027845 | - | 100 | 99.500 | Macaca_nemestrina |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSMLEG00000007082 | - | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSMLEG00000026260 | - | 93 | 97.500 | Mandrillus_leucophaeus |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSMAMG00000011635 | arl4aa | 100 | 82.000 | Mastacembelus_armatus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSMZEG00005017593 | - | 100 | 80.597 | Maylandia_zebra |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSMZEG00005016785 | - | 100 | 80.597 | Maylandia_zebra |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSMAUG00000013648 | Arl4a | 100 | 98.500 | Mesocricetus_auratus |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSMICG00000044178 | ARL4A | 100 | 99.000 | Microcebus_murinus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMOCG00000019911 | Arl4a | 100 | 99.500 | Microtus_ochrogaster |
| ENSG00000122644 | ARL4A | 97 | 80.412 | ENSMMOG00000002585 | arl4aa | 98 | 80.412 | Mola_mola |
| ENSG00000122644 | ARL4A | 100 | 84.577 | ENSMODG00000014219 | ARL4A | 100 | 84.577 | Monodelphis_domestica |
| ENSG00000122644 | ARL4A | 100 | 99.500 | MGP_CAROLIEiJ_G0017715 | Arl4a | 100 | 99.500 | Mus_caroli |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMUSG00000047446 | Arl4a | 100 | 99.500 | Mus_musculus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | MGP_PahariEiJ_G0029472 | Arl4a | 100 | 99.500 | Mus_pahari |
| ENSG00000122644 | ARL4A | 100 | 99.500 | MGP_SPRETEiJ_G0018557 | Arl4a | 100 | 99.500 | Mus_spretus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSMPUG00000019302 | ARL4A | 100 | 99.500 | Mustela_putorius_furo |
| ENSG00000122644 | ARL4A | 100 | 98.500 | ENSMLUG00000006004 | ARL4A | 100 | 98.500 | Myotis_lucifugus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSNGAG00000023900 | Arl4a | 100 | 99.500 | Nannospalax_galili |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSNBRG00000018555 | - | 100 | 80.100 | Neolamprologus_brichardi |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSNLEG00000016295 | - | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSODEG00000006469 | ARL4A | 100 | 98.000 | Octodon_degus |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSODEG00000019848 | - | 95 | 93.000 | Octodon_degus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSONIG00000020477 | arl4aa | 100 | 80.500 | Oreochromis_niloticus |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSONIG00000021291 | - | 100 | 81.592 | Oreochromis_niloticus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSOCUG00000000108 | ARL4A | 100 | 99.500 | Oryctolagus_cuniculus |
| ENSG00000122644 | ARL4A | 95 | 80.952 | ENSORLG00000029448 | arl4aa | 98 | 80.952 | Oryzias_latipes |
| ENSG00000122644 | ARL4A | 92 | 82.065 | ENSORLG00020011342 | arl4aa | 97 | 82.065 | Oryzias_latipes_hni |
| ENSG00000122644 | ARL4A | 96 | 80.729 | ENSORLG00015017167 | arl4aa | 98 | 80.729 | Oryzias_latipes_hsok |
| ENSG00000122644 | ARL4A | 92 | 81.967 | ENSOMEG00000003868 | arl4aa | 96 | 81.967 | Oryzias_melastigma |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSOGAG00000007283 | ARL4A | 100 | 99.500 | Otolemur_garnettii |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSOARG00000008124 | ARL4A | 100 | 99.500 | Ovis_aries |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSPPAG00000040138 | - | 90 | 91.053 | Pan_paniscus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSPPAG00000022226 | ARL4A | 100 | 99.500 | Pan_paniscus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSPPRG00000015025 | ARL4A | 100 | 99.500 | Panthera_pardus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSPTIG00000001360 | ARL4A | 100 | 99.500 | Panthera_tigris_altaica |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSPTRG00000043505 | - | 100 | 100.000 | Pan_troglodytes |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSPTRG00000048701 | ARL4A | 100 | 100.000 | Pan_troglodytes |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSPTRG00000052279 | - | 80 | 91.053 | Pan_troglodytes |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSPANG00000009329 | - | 100 | 99.000 | Papio_anubis |
| ENSG00000122644 | ARL4A | 100 | 100.000 | ENSPANG00000015865 | ARL4A | 100 | 100.000 | Papio_anubis |
| ENSG00000122644 | ARL4A | 100 | 80.500 | ENSPKIG00000010744 | arl4ab | 88 | 80.500 | Paramormyrops_kingsleyae |
| ENSG00000122644 | ARL4A | 100 | 81.500 | ENSPKIG00000011618 | arl4aa | 100 | 81.500 | Paramormyrops_kingsleyae |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSPSIG00000000003 | ARL4A | 100 | 96.000 | Pelodiscus_sinensis |
| ENSG00000122644 | ARL4A | 97 | 81.959 | ENSPMGG00000021471 | arl4aa | 99 | 81.959 | Periophthalmus_magnuspinnatus |
| ENSG00000122644 | ARL4A | 100 | 66.000 | ENSPMAG00000010027 | ARL4A | 100 | 64.500 | Petromyzon_marinus |
| ENSG00000122644 | ARL4A | 99 | 60.606 | ENSPMAG00000010406 | ARL4A | 99 | 60.606 | Petromyzon_marinus |
| ENSG00000122644 | ARL4A | 100 | 85.075 | ENSPCIG00000029346 | ARL4A | 100 | 85.075 | Phascolarctos_cinereus |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSPFOG00000020164 | arl4aa | 100 | 79.500 | Poecilia_formosa |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSPLAG00000017420 | arl4aa | 100 | 79.500 | Poecilia_latipinna |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSPPYG00000025824 | ARL4A | 100 | 99.500 | Pongo_abelii |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSPVAG00000005221 | - | 100 | 81.500 | Pteropus_vampyrus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSPNYG00000019193 | - | 100 | 80.597 | Pundamilia_nyererei |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSPNYG00000013877 | arl4aa | 100 | 81.000 | Pundamilia_nyererei |
| ENSG00000122644 | ARL4A | 95 | 78.421 | ENSPNAG00000009887 | arl4ab | 98 | 78.421 | Pygocentrus_nattereri |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSRNOG00000004282 | Arl4a | 100 | 99.500 | Rattus_norvegicus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSRBIG00000001927 | ARL4A | 100 | 99.500 | Rhinopithecus_bieti |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSRROG00000037376 | ARL4A | 100 | 99.500 | Rhinopithecus_roxellana |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSSBOG00000036387 | ARL4A | 100 | 99.500 | Saimiri_boliviensis_boliviensis |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSSFOG00015020603 | arl4aa | 100 | 84.000 | Scleropages_formosus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSSFOG00015015390 | ARL4A | 69 | 81.481 | Scleropages_formosus |
| ENSG00000122644 | ARL4A | 100 | 81.592 | ENSSMAG00000017183 | - | 99 | 81.592 | Scophthalmus_maximus |
| ENSG00000122644 | ARL4A | 100 | 82.000 | ENSSMAG00000015059 | arl4aa | 100 | 80.500 | Scophthalmus_maximus |
| ENSG00000122644 | ARL4A | 100 | 82.090 | ENSSDUG00000011151 | - | 100 | 82.090 | Seriola_dumerili |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSSDUG00000000586 | arl4aa | 100 | 81.000 | Seriola_dumerili |
| ENSG00000122644 | ARL4A | 91 | 83.516 | ENSSLDG00000012798 | - | 98 | 83.516 | Seriola_lalandi_dorsalis |
| ENSG00000122644 | ARL4A | 95 | 81.053 | ENSSLDG00000015190 | arl4aa | 97 | 81.053 | Seriola_lalandi_dorsalis |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSSARG00000003840 | ARL4A | 100 | 99.000 | Sorex_araneus |
| ENSG00000122644 | ARL4A | 100 | 95.500 | ENSSPUG00000008413 | ARL4A | 100 | 95.500 | Sphenodon_punctatus |
| ENSG00000122644 | ARL4A | 100 | 84.000 | ENSSPAG00000004205 | - | 100 | 82.673 | Stegastes_partitus |
| ENSG00000122644 | ARL4A | 94 | 81.818 | ENSSPAG00000008565 | arl4aa | 97 | 81.818 | Stegastes_partitus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSSSCG00000032709 | ARL4A | 100 | 99.500 | Sus_scrofa |
| ENSG00000122644 | ARL4A | 100 | 97.500 | ENSTGUG00000002488 | ARL4A | 100 | 97.500 | Taeniopygia_guttata |
| ENSG00000122644 | ARL4A | 100 | 80.392 | ENSTRUG00000008027 | arl4aa | 100 | 75.253 | Takifugu_rubripes |
| ENSG00000122644 | ARL4A | 100 | 78.431 | ENSTNIG00000000601 | arl4aa | 100 | 74.627 | Tetraodon_nigroviridis |
| ENSG00000122644 | ARL4A | 100 | 96.000 | ENSTBEG00000004057 | - | 97 | 90.909 | Tupaia_belangeri |
| ENSG00000122644 | ARL4A | 100 | 99.000 | ENSTTRG00000012582 | ARL4A | 100 | 99.000 | Tursiops_truncatus |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSUAMG00000009218 | ARL4A | 100 | 99.500 | Ursus_americanus |
| ENSG00000122644 | ARL4A | 100 | 98.000 | ENSVPAG00000002002 | ARL4A | 100 | 98.000 | Vicugna_pacos |
| ENSG00000122644 | ARL4A | 100 | 99.500 | ENSVVUG00000022573 | ARL4A | 100 | 99.500 | Vulpes_vulpes |
| ENSG00000122644 | ARL4A | 100 | 89.000 | ENSXETG00000011342 | arl4a | 100 | 89.000 | Xenopus_tropicalis |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSXCOG00000007236 | arl4aa | 100 | 79.500 | Xiphophorus_couchianus |
| ENSG00000122644 | ARL4A | 100 | 80.000 | ENSXMAG00000019995 | arl4aa | 100 | 79.500 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 10980193.17398095.25416956. | IPI | Function |
| GO:0005525 | GTP binding | 21873635. | IBA | Function |
| GO:0005525 | GTP binding | 10980193. | IDA | Function |
| GO:0005634 | nucleus | 10980193. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005730 | nucleolus | 10980193. | IDA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005886 | plasma membrane | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | 17398095. | IDA | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0016192 | vesicle-mediated transport | 21873635. | IBA | Process |
| GO:0050873 | brown fat cell differentiation | - | IEA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| LUAD | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| KIRP | ||
| PCPG | ||
| BLCA | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| THCA | ||
| LUAD |