
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 14647465 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000609821 | PLCG1-218 | 570 | - | - | - (aa) | - | - |
| ENST00000477870 | PLCG1-208 | 1051 | - | - | - (aa) | - | - |
| ENST00000617873 | PLCG1-220 | 583 | - | - | - (aa) | - | - |
| ENST00000609257 | PLCG1-217 | 956 | - | ENSP00000476757 | 133 (aa) | - | V9GYH5 |
| ENST00000599785 | PLCG1-213 | 880 | - | ENSP00000468958 | 50 (aa) | - | M0QX77 |
| ENST00000630423 | PLCG1-222 | 635 | - | - | - (aa) | - | - |
| ENST00000244007 | PLCG1-201 | 5285 | XM_005260438 | ENSP00000244007 | 1291 (aa) | XP_005260495 | P19174 |
| ENST00000483646 | PLCG1-210 | 591 | - | - | - (aa) | - | - |
| ENST00000483175 | PLCG1-209 | 1084 | - | - | - (aa) | - | - |
| ENST00000465571 | PLCG1-205 | 543 | - | - | - (aa) | - | - |
| ENST00000423733 | PLCG1-203 | 498 | - | - | - (aa) | - | - |
| ENST00000373271 | PLCG1-202 | 7395 | XM_011528867 | ENSP00000362368 | 1290 (aa) | XP_011527169 | P19174 |
| ENST00000490253 | PLCG1-211 | 527 | - | - | - (aa) | - | - |
| ENST00000619272 | PLCG1-221 | 330 | - | - | - (aa) | - | - |
| ENST00000470528 | PLCG1-206 | 510 | - | - | - (aa) | - | - |
| ENST00000608885 | PLCG1-216 | 865 | - | ENSP00000476432 | 219 (aa) | - | V9GY63 |
| ENST00000461641 | PLCG1-204 | 936 | - | - | - (aa) | - | - |
| ENST00000492148 | PLCG1-212 | 597 | - | - | - (aa) | - | - |
| ENST00000473632 | PLCG1-207 | 412 | - | - | - (aa) | - | - |
| ENST00000608689 | PLCG1-215 | 969 | - | - | - (aa) | - | - |
| ENST00000607954 | PLCG1-214 | 1005 | - | ENSP00000476454 | 108 (aa) | - | V9GY71 |
| ENST00000612731 | PLCG1-219 | 544 | - | ENSP00000485760 | 132 (aa) | - | A0A0D9SEK2 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00562 | Inositol phosphate metabolism | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | KEGG |
| hsa04012 | ErbB signaling pathway | KEGG |
| hsa04014 | Ras signaling pathway | KEGG |
| hsa04015 | Rap1 signaling pathway | KEGG |
| hsa04020 | Calcium signaling pathway | KEGG |
| hsa04064 | NF-kappa B signaling pathway | KEGG |
| hsa04066 | HIF-1 signaling pathway | KEGG |
| hsa04070 | Phosphatidylinositol signaling system | KEGG |
| hsa04072 | Phospholipase D signaling pathway | KEGG |
| hsa04360 | Axon guidance | KEGG |
| hsa04370 | VEGF signaling pathway | KEGG |
| hsa04650 | Natural killer cell mediated cytotoxicity | KEGG |
| hsa04658 | Th1 and Th2 cell differentiation | KEGG |
| hsa04659 | Th17 cell differentiation | KEGG |
| hsa04660 | T cell receptor signaling pathway | KEGG |
| hsa04664 | Fc epsilon RI signaling pathway | KEGG |
| hsa04666 | Fc gamma R-mediated phagocytosis | KEGG |
| hsa04670 | Leukocyte transendothelial migration | KEGG |
| hsa04722 | Neurotrophin signaling pathway | KEGG |
| hsa04750 | Inflammatory mediator regulation of TRP channels | KEGG |
| hsa04919 | Thyroid hormone signaling pathway | KEGG |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | KEGG |
| hsa05110 | Vibrio cholerae infection | KEGG |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | KEGG |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | KEGG |
| hsa05170 | Human immunodeficiency virus 1 infection | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| hsa05205 | Proteoglycans in cancer | KEGG |
| hsa05206 | MicroRNAs in cancer | KEGG |
| hsa05214 | Glioma | KEGG |
| hsa05223 | Non-small cell lung cancer | KEGG |
| hsa05225 | Hepatocellular carcinoma | KEGG |
| hsa05231 | Choline metabolism in cancer | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000124181 | Blood Pressure | 4E-8 | 27790247 |
| ENSG00000124181 | Cholesterol | 4E-8 | 27790247 |
| ENSG00000124181 | Electrocardiography | 4E-8 | 27790247 |
| ENSG00000124181 | Glucose | 4E-8 | 27790247 |
| ENSG00000124181 | Hematocrit | 4E-8 | 27790247 |
| ENSG00000124181 | Cholesterol, HDL | 4E-8 | 27790247 |
| ENSG00000124181 | Triglycerides | 4E-8 | 27790247 |
| ENSG00000124181 | Body Mass Index | 4E-8 | 27790247 |
| ENSG00000124181 | Atrial Fibrillation | 7E-7 | 27790247 |
| ENSG00000124181 | Blood Pressure | 7E-7 | 27790247 |
| ENSG00000124181 | Cholesterol | 7E-7 | 27790247 |
| ENSG00000124181 | Coronary Disease | 7E-7 | 27790247 |
| ENSG00000124181 | Diabetes Mellitus | 7E-7 | 27790247 |
| ENSG00000124181 | Electrocardiography | 7E-7 | 27790247 |
| ENSG00000124181 | Glucose | 7E-7 | 27790247 |
| ENSG00000124181 | Heart Failure | 7E-7 | 27790247 |
| ENSG00000124181 | Hematocrit | 7E-7 | 27790247 |
| ENSG00000124181 | Cholesterol, HDL | 7E-7 | 27790247 |
| ENSG00000124181 | Mortality | 7E-7 | 27790247 |
| ENSG00000124181 | Neoplasms | 7E-7 | 27790247 |
| ENSG00000124181 | Triglycerides | 7E-7 | 27790247 |
| ENSG00000124181 | Body Mass Index | 7E-7 | 27790247 |
| ENSG00000124181 | Stroke | 7E-7 | 27790247 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124181 | rs2228246 | 20 | 41163423 | G | Major depressive disorder | 27479909 | [0.026-0.058] unit increase | 0.042 | EFO_0003761 |
| ENSG00000124181 | rs760762 | 20 | 41147406 | ? | Age-related disease endophenotypes | 27790247 | EFO_0004574|EFO_0006336|EFO_0004530|EFO_0006335|EFO_0004348|EFO_0007928|EFO_0004468|EFO_0004340|EFO_0004612 | ||
| ENSG00000124181 | rs6129767 | 20 | 41193692 | T | Coronary artery disease | 29212778 | [0.025-0.052] unit decrease | 0.0385 | EFO_0000378 |
| ENSG00000124181 | rs753381 | 20 | 41168825 | C | Hemoglobin concentration | 27863252 | [0.014-0.028] unit increase | 0.02054587 | EFO_0004509 |
| ENSG00000124181 | rs4297946 | 20 | 41182635 | G | Total cholesterol levels | 29507422 | unit decrease | 0.029 | EFO_0004574 |
| ENSG00000124181 | rs4297946 | 20 | 41182635 | G | Total cholesterol levels | 29507422 | unit decrease | 0.027 | EFO_0004574 |
| ENSG00000124181 | rs4297946 | 20 | 41182635 | G | Low density lipoprotein cholesterol levels | 29507422 | unit decrease | 0.032 | EFO_0004611 |
| ENSG00000124181 | rs4297946 | 20 | 41182635 | G | Low density lipoprotein cholesterol levels | 29507422 | unit decrease | 0.03 | EFO_0004611 |
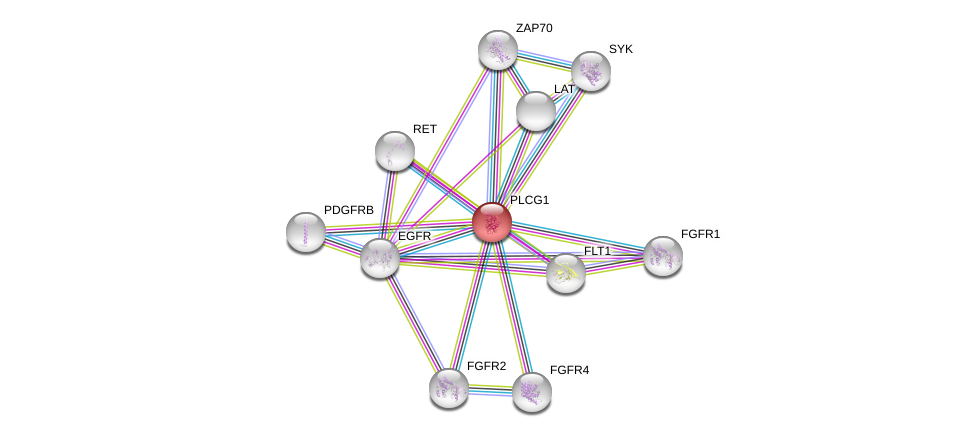
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001701 | in utero embryonic development | - | IEA | Process |
| GO:0001726 | ruffle | 17229814. | IDA | Component |
| GO:0004435 | phosphatidylinositol phospholipase C activity | 21873635. | IBA | Function |
| GO:0004435 | phosphatidylinositol phospholipase C activity | 2550068. | IDA | Function |
| GO:0004435 | phosphatidylinositol phospholipase C activity | - | TAS | Function |
| GO:0004629 | phospholipase C activity | - | TAS | Function |
| GO:0005168 | neurotrophin TRKA receptor binding | 15488758. | IPI | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 1396585.1656221.7537096.7537265.7693694.7993895.8321198.8627166.8628282.8657103.8892607.9020117.9045636.9489702.9593725.10811803.10873601.11796522.11896612.14568990.14570588.14722116.15641795.15929943.16273093.16525023.16921170.16938345.17229814.17474147.17936057.19264842.20808760.20932831.21706016.21725281.23063561.23397142.23460737.24440983.24642916.24728074.25241761. | IPI | Function |
| GO:0005737 | cytoplasm | 15702972. | IDA | Component |
| GO:0005829 | cytosol | 2550068. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | 2550068.22454520. | IDA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005911 | cell-cell junction | - | IEA | Component |
| GO:0007165 | signal transduction | - | TAS | Process |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 22454520. | IMP | Process |
| GO:0007202 | activation of phospholipase C activity | - | TAS | Process |
| GO:0007411 | axon guidance | - | TAS | Process |
| GO:0008180 | COP9 signalosome | 22561606. | IDA | Component |
| GO:0009395 | phospholipid catabolic process | - | IEA | Process |
| GO:0010634 | positive regulation of epithelial cell migration | 21873635. | IBA | Process |
| GO:0010634 | positive regulation of epithelial cell migration | 17229814. | IMP | Process |
| GO:0010863 | positive regulation of phospholipase C activity | - | TAS | Process |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016477 | cell migration | 22454520. | IMP | Process |
| GO:0019722 | calcium-mediated signaling | 22454520. | IMP | Process |
| GO:0019901 | protein kinase binding | 22454520. | IPI | Function |
| GO:0030027 | lamellipodium | 17229814. | IDA | Component |
| GO:0030971 | receptor tyrosine kinase binding | - | IEA | Function |
| GO:0032959 | inositol trisphosphate biosynthetic process | 21873635. | IBA | Process |
| GO:0035254 | glutamate receptor binding | - | IEA | Function |
| GO:0038095 | Fc-epsilon receptor signaling pathway | - | TAS | Process |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | - | TAS | Process |
| GO:0042995 | cell projection | 22454520. | IDA | Component |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | 20011604. | IDA | Process |
| GO:0043647 | inositol phosphate metabolic process | - | TAS | Process |
| GO:0045766 | positive regulation of angiogenesis | 20011604. | IDA | Process |
| GO:0048015 | phosphatidylinositol-mediated signaling | 21873635. | IBA | Process |
| GO:0050804 | modulation of chemical synaptic transmission | - | IEA | Process |
| GO:0050852 | T cell receptor signaling pathway | - | TAS | Process |
| GO:0050900 | leukocyte migration | - | TAS | Process |
| GO:0051209 | release of sequestered calcium ion into cytosol | 21873635. | IBA | Process |
| GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 22454520. | IMP | Process |
| GO:0071364 | cellular response to epidermal growth factor stimulus | 17229814. | IDA | Process |
| GO:0071364 | cellular response to epidermal growth factor stimulus | 22454520. | IMP | Process |
| GO:0098685 | Schaffer collateral - CA1 synapse | - | IEA | Component |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:1905564 | positive regulation of vascular endothelial cell proliferation | 27464494. | IMP | Process |
| GO:2000353 | positive regulation of endothelial cell apoptotic process | 27464494. | IMP | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| COAD | |||
| LAML | |||
| LUAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| PAAD | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| THCA | ||
| LUAD | ||
| UVM |