
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000265462 | PRDX5-201 | 893 | - | ENSP00000265462 | 214 (aa) | - | P30044 |
| ENST00000352435 | PRDX5-203 | 596 | - | ENSP00000335334 | 170 (aa) | - | P30044 |
| ENST00000347941 | PRDX5-202 | 463 | - | ENSP00000335363 | 125 (aa) | - | P30044 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04146 | Peroxisome | KEGG |
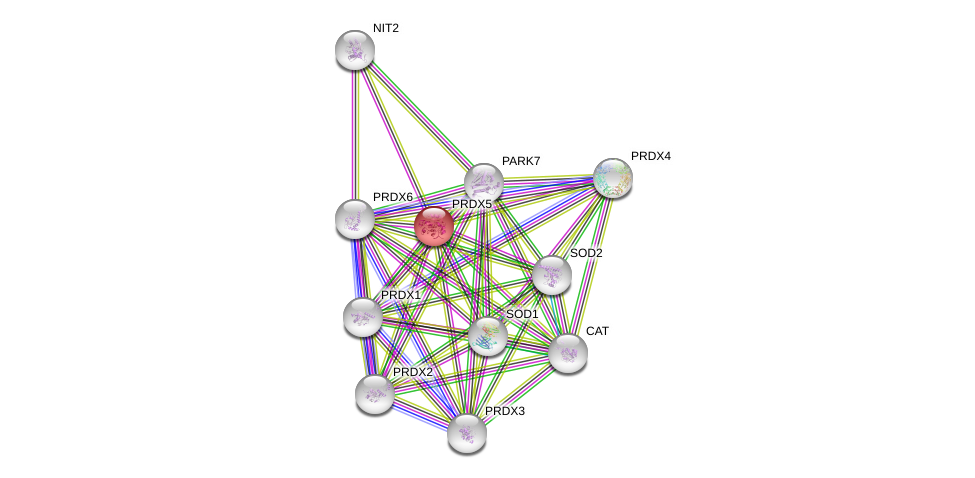
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001016 | RNA polymerase III regulatory region DNA binding | 10095767. | IDA | Function |
| GO:0004601 | peroxidase activity | 10521424. | IDA | Function |
| GO:0005102 | signaling receptor binding | 20178365. | IPI | Function |
| GO:0005515 | protein binding | 29128334. | IPI | Function |
| GO:0005615 | extracellular space | 23580065. | HDA | Component |
| GO:0005634 | nucleus | 10095767.15785239. | IDA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 15785239. | IDA | Component |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 10521424.10751410.18262354. | IDA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0005777 | peroxisome | 21873635. | IBA | Component |
| GO:0005777 | peroxisome | 10521424. | IDA | Component |
| GO:0005782 | peroxisomal matrix | - | IEA | Component |
| GO:0005829 | cytosol | 18262354. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006915 | apoptotic process | - | IEA | Process |
| GO:0006954 | inflammatory response | 10521424. | TAS | Process |
| GO:0006979 | response to oxidative stress | 10521424. | IDA | Process |
| GO:0008379 | thioredoxin peroxidase activity | 15280035. | EXP | Function |
| GO:0008379 | thioredoxin peroxidase activity | 21873635. | IBA | Function |
| GO:0016480 | negative regulation of transcription by RNA polymerase III | 10095767. | IDA | Process |
| GO:0031410 | cytoplasmic vesicle | 10095767. | IDA | Component |
| GO:0034599 | cellular response to oxidative stress | 21873635. | IBA | Process |
| GO:0034599 | cellular response to oxidative stress | - | TAS | Process |
| GO:0034614 | cellular response to reactive oxygen species | 18262354. | IMP | Process |
| GO:0042744 | hydrogen peroxide catabolic process | 21873635. | IBA | Process |
| GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process | 18262354. | IMP | Function |
| GO:0043066 | negative regulation of apoptotic process | 18262354. | IMP | Process |
| GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | - | IEA | Process |
| GO:0043231 | intracellular membrane-bounded organelle | 15785239. | IDA | Component |
| GO:0045454 | cell redox homeostasis | 21873635. | IBA | Process |
| GO:0046983 | protein dimerization activity | 15046979. | IDA | Function |
| GO:0048471 | perinuclear region of cytoplasm | 10095767. | IDA | Component |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.23533145. | HDA | Component |
| GO:0098869 | cellular oxidant detoxification | - | IEA | Process |